Data Engineering Zoomcamp FAQ
Table of Contents
General Course-Related Questions
# Course: When does the course start?
The next cohort starts January 13th, 2025. More info at DTC.
- Register before the course starts using this link.
- Join the course Telegram channel with announcements.
- Don’t forget to register in DataTalks.Club's Slack and join the channel.
# Course: What are the prerequisites for this course?
To get the most out of this course, you should have:
- Basic coding experience
- Familiarity with SQL
- Experience with Python (helpful but not required)
No prior data engineering experience is necessary. See Readme on GitHub.
# Course: Can I still join the course after the start date?
Yes, even if you don't register, you're still eligible to submit the homework.
Be aware, however, that there will be deadlines for turning in homeworks and the final projects. So don't leave everything for the last minute.
# Course: I have registered for the Data Engineering Bootcamp. When can I expect to receive the confirmation email?
You don't need a confirmation email. You're accepted. You can start learning and submitting homework without registering. Registration was just to gauge interest before the start date.
# Course: What can I do before the course starts?
Start by installing and setting up all the dependencies and requirements:
- Google Cloud account
- Google Cloud SDK
- Python 3 (installed with Anaconda)
- Terraform
- Git
Look over the prerequisites and syllabus to see if you are comfortable with these subjects.
# Course: how many Zoomcamps in a year?
There are multiple Zoomcamps in a year, as of 2025. More info at DTC Article.
They are five separate courses, estimated to be during these months:
- Data-Engineering: Jan - Apr
- Stock Market Analytics: Apr - May
- MLOps: May - Aug
- LLM: June - Sep
- Machine Learning: Sep - Jan
There's only one Data-Engineering Zoomcamp “live” cohort per year for the certification, similar to the other Zoomcamps. They follow pretty much the same schedule for each cohort. For Data-Engineering, it is generally from Jan-Apr of the year.
If you’re not interested in the Certificate, you can take any Zoomcamp at any time, at your own pace, out of sync with any “live” cohort.
# Course: Is the current cohort going to be different from the previous cohort?
For the 2025 edition, we are using Kestra (see Demo) instead of MageAI (Module 2). Look out for new videos. See Playlist.
For the 2024 edition, we used Mage AI instead of Prefect and re-recorded the Terraform videos. For 2023, we used Prefect instead of Airflow. See playlists on YouTube and the cohorts folder in the GitHub repo.
# Course - Can I follow the course after it finishes?
Yes, we will keep all the materials available, so you can follow the course at your own pace after it finishes.
You can also continue reviewing the homeworks and prepare for the next cohort. You can also start working on your final capstone project.
# Course: Can I get support if I take the course in the self-paced mode?
Yes, the Slack channel remains open and you can ask questions there. However, always search the channel first and check the FAQ, as most likely your questions are already answered here.
You can also tag the bot @ZoomcampQABot to help you conduct the search, but don’t rely on its answers 100%.
# Course: Which playlist on YouTube should I refer to?
All the main videos are stored in the "DATA ENGINEERING ZOOMCAMP" main playlist. The GitHub repository is updated to include each video with a thumbnail, linking directly to the relevant playlist.
Refer to the Main Playlist for the core content, and then check specific year playlists for additional videos such as office hours.
# Course: How many hours per week am I expected to spend on this course?
It depends on your background and previous experience with modules. It is expected to require about 5 - 15 hours per week.
You can also calculate it yourself using this data and then update this answer.
# Office Hours: I can’t attend the “Office hours” / workshop, will it be recorded?
Yes! Every "Office Hours" will be recorded and available a few minutes after the live session is over; so you can view (or rewatch) whenever you want.
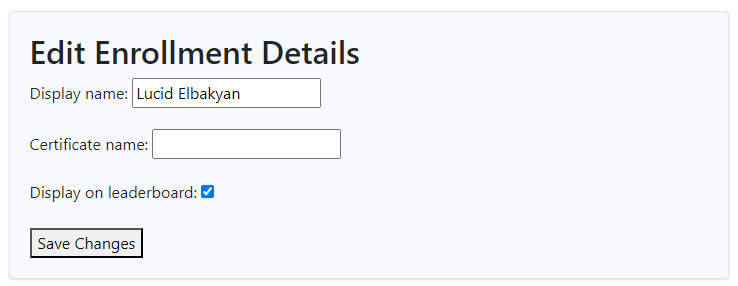
# Edit Course Profile.
The display name listed on the leaderboard is an auto-generated randomized name. You can edit it to be a nickname or your real name if you prefer. Your entry on the Leaderboard is the one highlighted in light green.
The Certificate name should be your actual name that you want to appear on your certificate after completing the course.
The "Display on Leaderboard" option indicates whether you want your name to be listed on the course leaderboard.
# Certificate: Do I need to do the homeworks to get the certificate?
No, as long as you complete the peer-reviewed capstone projects on time, you can receive the certificate. You do not need to do the homeworks if you join late, for example.
# Certificate - Can I follow the course in a self-paced mode and get a certificate?
No, you can only get a certificate if you finish the course with a “live” cohort. We don't award certificates for the self-paced mode. The reason is you need to peer-review capstone(s) after submitting a project. You can only peer-review projects at the time the course is running.
# Homework: What are homework and project deadlines?
2025 deadlines will be announced on the course website and in Google Calendar.
You can find the 2024 deadlines here: 2024 Deadlines Spreadsheet.
Also, take note of announcements from @Au-Tomator for any extensions or other news. The form may also show the updated deadline if the instructor(s) have updated it.
# Homework: Are late submissions of homework allowed?
No, late submissions are not allowed. However, if the form is still open after the due date, you can still submit the homework. Confirm your submission by checking the date-timestamp on the Course page. Ensure you are logged in.
# Homework and Leaderboard: What is the system for points in the course management
After you submit your homework, it will be graded based on the number of questions in that particular assignment. You can see the number of points you have earned at the top of the homework page. Additionally, in the leaderboard, you will find the sum of all points you've earned: points for Homeworks, FAQs, and Learning in Public.
Point System Overview:
- Homework: Points vary by assignment based on the number of questions.
- FAQ Contribution: You get a maximum of 1 point for contributing to the FAQ in the respective week.
- Learning in Public: For each learning in public link, you earn one point. You can achieve a maximum of 7 points.
Check this video for more details.
# Homework: What is the homework URL in the homework link?

Answer: In short, it’s your repository on GitHub, GitLab, Bitbucket, etc.
In long, your repository or any other location where you have your code, and a reasonable person would look at it and think, yes, you went through the week and exercises. Think of it like a portfolio you could present to an employer.
# Leaderboard: how do find myself on the leaderboard?
When you set up your account, you are automatically assigned a random name, such as "Lucid Elbakyan." If you want to see what your display name is, follow these steps:
- Go to your profile.
- Log in.
- Your display name is shown. You can also change it if you wish.
- Ensure your certificate name is correct, as this name will later be printed on your certificate.
# Environment: Is Python 3.9 still the recommended version to use in 2024?
Yes, for simplicity and stability when troubleshooting against recorded videos.
But Python 3.10 and 3.11 should work fine.
# Environment - Should I use my local machine, GCP, or GitHub Codespaces for my environment?
You can set it up on your laptop or PC if you prefer to work locally. However, Windows users might face some challenges.
If you prefer to work on the local machine, you can start with the Week 1 Introduction to Docker.
Alternatively, if you prefer to set up a virtual machine, consider the following:
- Using GitHub Codespaces
- Setting up the environment on a cloud VM: Refer to this video for guidance.
Working on a virtual machine is beneficial if you have different devices for home and office, allowing you to work virtually anywhere.
# Environment - Is GitHub codespaces an alternative to using cli/git bash to ingest the data and create a docker file?
GitHub Codespaces offers you computing Linux resources with many pre-installed tools (Docker, Docker Compose, Python).
You can also open any GitHub repository in a GitHub Codespace.
# Environment: Do we really have to use GitHub codespaces? I already have PostgreSQL & Docker installed.
It's up to you which platform and environment you use for the course.
GitHub Codespaces or GCP VM are just possible options, but you can do the entire course from your laptop.
# Environment - Do I need both GitHub Codespaces and GCP?
Choose the approach that aligns the most with your idea for the end project.
One should suffice; however, BigQuery, which is part of GCP, will be used, so learning that is probably a better option. Alternatively, you can set up a local environment for most of this course.
# Environment - Could not establish connection to "MyServerName": Got bad result from install script
This issue occurs when attempting to connect to a GCP VM using VSCode on a Windows machine. You can resolve it by changing a registry value in the registry editor.
Open the Run command window:
- Use the shortcut keys
Windows + R, or - Right-click "Start" and click "Run".
Open the Registry Editor:
- Type
regeditin the Run command window, then press Enter.
Change the registry value:
- Navigate to
HKEY_CURRENT_USER\Software\Microsoft\Command Processor. - Change the "Autorun" value from "if exists" to a blank.
Alternatively, you can delete the saved fingerprint within the known_hosts file:
In Windows, locate the file at C:\Users\<your_user_name>\.ssh\known_hosts and remove the entry for the server.
# Environment - Why are we using GCP and not other cloud providers?
For uniformity.
You can use other cloud platforms since you get every service provided by GCP in Azure and AWS. You’re not restricted to GCP and can use other platforms like AWS if you’re more comfortable.
Because everyone usually has a Google account, GCP offers a free trial period with $300 in credits for new users. Additionally, we are working with BigQuery, which is part of GCP.
Note that to sign up for a free GCP account, you must have a valid credit card.
# Should I pay for cloud services?
It's not mandatory. You can take advantage of their free trial.
# Environment: The GCP and other cloud providers are unavailable in some countries. Is it possible to provide a guide to installing a home lab?
You can do most of the course without a cloud. Almost everything we use (excluding BigQuery) can be run locally. We won’t be able to provide guidelines for some things, but most of the materials are runnable without GCP.
For everything in the course, there’s a local alternative. You could even do the whole course locally. Note that Homework 3 requires BigQuery.
# Environment: Is GCP Sandbox enough or we need the Free Trial?
Google Cloud Platform (GCP) provides two free trial options: the Free Trial with $300 credit and the Sandbox. Users can switch between these options by managing billing details.
However, completing the course solely using the GCP Sandbox option is not feasible due to its limited features. The Sandbox lacks some services required for the course, such as VMs, GCS Buckets, and other paid services that are integral to the curriculum.
The course will eventually require utilizing the following:
- VMs and GCS Buckets: These resources are not fully available in the Sandbox.
- BigQuery: A key component of GCP, and the Sandbox may not support all necessary functionalities.
Therefore, it is recommended to use the GCP Free Trial with billing details to access all needed features and ensure a smooth learning experience.
# Environment: I want to use AWS. May I do that?
Yes, you can. Just remember to adapt all the information from the videos to AWS. Additionally, the final capstone will be evaluated based on these tasks:
- Create a data pipeline
- Develop a visualization
Consider that when seeking help, you might need to rely on fellow coursemates who use AWS, which could be fewer compared to those using GCP.
Also, see "Is it possible to use x tool instead of the one tool you use?"
# Besides the “Office Hour” which are the live zoom calls?
We will probably have some calls during the Capstone period to clear some questions, but it will be announced in advance if that happens.
See Google Calendar
# Can I use Airflow instead for my final project?
Yes, you can use any tool you want for your project.
# Is it possible to use tool “X” instead of the one tool you use in the course?
Yes, this applies if you want to use Airflow or Prefect instead of Mage, AWS or Snowflake instead of GCP products, or Tableau instead of Metabase or Google Data Studio.
The course covers two alternative data stacks: one using GCP and one using local installation of everything. You can use one of them or use your tool of choice.
Considerations:
- We can’t support you if you choose to use a different stack.
- You would need to explain the different choices of tools for the peer review of your capstone project.
# How can we contribute to the course?
- Star the repository.
- Share it with friends if you find it useful.
- Create a pull request (PR) if you can improve the text or structure of the repository.
- Update this FAQ.
# Environment: Is the course [Windows/macOS/Linux/...] friendly?
Yes! Linux is ideal but technically it should not matter. Students in the 2024 cohort used all 3 OSes successfully.
# Environment: Roadblock for Windows users in modules with *.sh (shell scripts)
Later modules (module-05 & RisingWave workshop) use shell scripts in *.sh files. Most Windows users not using WSL will encounter issues and may not be able to continue, even in Git Bash or MINGW64. It is recommended to set up a WSL environment from the start.
# Any books or additional resources you recommend?
Yes to both! Check out this document: Awesome Data Engineering Resources
# Project: What is Project Attempt #1 and Project Attempt #2 exactly?
You will have two attempts for a project.
- If the first project deadline is over and you’re late, or you submit the project and fail the first attempt, you have another chance to submit the project with the second attempt.
# How to troubleshoot issues
First Steps:
- Attempt to solve the issue independently. Familiarize yourself with documentation as a crucial skill for problem-solving.
- Use shortcuts like
[ctrl+f]to search within documents and browsers. - Analyze the error message for descriptions, instructions, and possible solutions.
- Restart your application, server, or computer as needed.
Search for Solutions:
- Use search engines like Google, ChatGPT, or Bing AI to research the issue. It is rare to encounter a unique problem.
- Form your search queries using:
<technology> <problem statement>. E.g.,pgcli error column c.relhasoids does not exist. - Consult the technology’s official documentation for guidance.
Uninstallation and Reinstallation:
- Uninstall and then reinstall the application if needed, including a system restart.
- Note that reinstalling without prior uninstallation might not resolve the issue.
Seeking Help:
- Post questions on platforms like StackOverflow. Ensure your question adheres to guidelines: how-to-ask.
- Consider asking experts or colleagues in the future.
Community Resources:
- Check Slack channels for pinned messages and use its search function.
- Refer to this FAQ using search
[ctrl+f]or utilize the@ZoomcampQABotfor assistance.
When Asking for Help:
- Provide detailed information: coding environment, OS, commands, videos followed, etc.
- Share errors received, with specifics, including line numbers and actions taken.
- Avoid screenshots; paste code or errors directly. Use ``` for code formatting.
- Maintain thread consistency; respond in the same thread instead of creating multiple ones.
Re-evaluation:
- If the issue recurs, create a new post detailing changes in the environment.
- Communicate additional troubleshooting steps in the same thread.
- Occasionally take a break to gain a fresh perspective on the problem.
Documentation Contribution:
- If your problem solution is not listed, consider adding it to the FAQ to assist others.
# How to ask questions
When the troubleshooting guide does not help resolve your issue and you need another pair of eyes, include as much information as possible when asking a question:
- What are you coding on? What operating system are you using?
- What command did you run, and which video or tutorial did you follow?
- What error did you get? Does it have a line number pointing to the problematic code, and have you checked it for typos?
- What have you tried that did not work? This is crucial because, without it, helpers might suggest steps mentioned in the error log first. Or just refer to this FAQ document.
# How do I use Git / GitHub for this course?
After you create a GitHub account, clone the course repo to your local machine using the process outlined in this video:
Git for Everybody: How to Clone a Repository from GitHub.
Having this local repository on your computer will make it easy to access the instructors’ code and make pull requests if you want to add your own notes or make changes to the course content.
You will probably also create your own repositories to host your notes and versions of files. Here is a great tutorial that shows you how to do this:
How to Create a Git Repository.
Remember to ignore large databases, .csv, and .gz files, and other files that should not be saved to a repository. Use .gitignore for this:
Important:
NEVER store passwords or keys in a git repo (even if the repo is set to private). Put files containing sensitive information (.env, secret.json, etc.) in your .gitignore.
This is also a great resource: Dangit, Git!?!
# VS Code: Tab using spaces
Error:
Makefile:2: *** missing separator. Stop.
Solution:
Tabs in documents should be converted to Tab instead of spaces. Follow this stack.
# Opening an HTML file with a Windows browser from Linux running on WSL
If you’re running Linux on Windows Subsystem for Linux (WSL) 2, you can open HTML files from the guest (Linux) with any Internet Browser installed on the host (Windows). Just install wslu and open the page using wslview:
wslview index.html
You can customize which browser to use by setting the BROWSER environment variable first. For example:
export BROWSER='/mnt/c/Program Files/Firefox/firefox.exe'
# Set up Chrome Remote Desktop for Linux on Compute Engine
This tutorial shows you how to set up the Chrome Remote Desktop service on a Debian Linux virtual machine (VM) instance on Compute Engine. Chrome Remote Desktop allows you to remotely access applications with a graphical user interface.
# How do I get my certificate?
There'll be an announcement in Telegram and the course channel for:
- Checking that your full name is displayed correctly on the Certificate (see Editing course profile on the Course Management webpage).
- Notifying when the grading is completed.
You will find it in your course profile (you need to be logged it).
For 2025 the link to the course profile is this:
https://courses.datatalks.club/de-zoomcamp-2025/enrollment
For other editions, change "2025" to your edition.
After the second announcement, follow instructions in certificates.md on how to generate the Certificate document yourself.
Module 1: Docker and Terraform
# Taxi Data: Yellow Taxi Trip Records downloading error
When attempting to download the 2021 data from the TLC website, you may encounter the following error:
ERROR 403: Forbidden

We have a backup, so use it instead: nyc-tlc-data
So the link should be yellow_tripdata_2021-01.csv.gz.
Note: Make sure to unzip the "gz" file (no, the "unzip" command won’t work for this).
# Taxi Data: How to handle *.csv.gz taxi data files?
In this video, the data file is stored as output.csv. If the file extension is csv.gz instead of csv, it won't store correctly.
To handle this:
Replace
csv_name = "output.csv"with the file name extracted from the URL. For example, for the yellow taxi data, use:url = "https://github.com/DataTalksClub/nyc-tlc-data/releases/download/yellow/yellow_tripdata_2021-01.csv.gz" csv_name = url.split("/")[-1]When you use
csv_namewithpandas.read_csv, it will work correctly becausepandas.read_csvcan directly read files with thecsv.gzextension.
Example:
import pandas as pd
url = "https://github.com/DataTalksClub/nyc-tlc-data/releases/download/yellow/yellow_tripdata_2021-01.csv.gz"
csv_name = url.split("/")[-1]
data = pd.read_csv(csv_name)
# Taxi Data: Data Dictionary for NY Taxi data?
Yellow Trips: Data Dictionary
Green Trips: Data Dictionary - LPEP Trip Records May 1, 2018
# Taxi Data: Unzip Parquet file
You can unzip the downloaded parquet file from the command line. The result is a CSV file which can be imported with pandas using pd.read_csv() as shown in the videos.
gunzip green_tripdata_2019-09.csv.gz
Solution for Using Parquet Files Directly in Python Script ingest_data.py
In the
def main(params), add this line:parquet_name = 'output.parquet'Edit the code which downloads the files:
os.system(f"wget {url} -O {parquet_name}")Convert the downloaded
.parquetfile to CSV and rename it tocsv_nameto keep it relevant to the rest of the code:df = pd.read_parquet(parquet_name) df.to_csv(csv_name, index=False)
# wget is not recognized as an internal or external command
If you encounter the error "wget is not recognized as an internal or external command," wget needs to be installed.
This error may also cause messages like "No such file or directory: 'output.csv.gz'."
Installation Instructions:
On Ubuntu:
sudo apt-get install wgetOn macOS:
Use Homebrew:
brew install wgetOn Windows:
Use Chocolatey:
choco install wgetAlternatively, download a binary from GnuWin32 and place it in a location that is in your PATH (e.g.,
C:/tools/).
Alternative Windows Installation:
- Download the latest wget binary for Windows from eternallybored.
- If you downloaded the zip, extract all files (use 7-zip if the built-in utility gives an error).
- Rename the file
wget64.exetowget.exeif necessary. - Move
wget.exeto yourGit\mingw64\bin\directory.
Python Alternative:
Use the Python wget library:
First, install using pip:
pip install wgetUse it with Python:
python -m wget
You can also paste the file URL into your web browser to download normally, then move the file to your working directory.
Additional Recommendation:
Consider using the Python library requests for loading gz files.
# wget - ERROR: cannot verify <website> certificate (MacOS)
Firstly, make sure that you add ! before wget if you’re running your command in a Jupyter Notebook or CLI. Then, you can check one of these two things (from CLI):
Using the Python library wget installed with pip:
python -m wget <url>Use the usual command and add
--no-check-certificateat the end:!wget <website_url> --no-check-certificate
# Git Bash: Backslash as an escape character in Git Bash for Windows
For those who wish to use the backslash as an escape character in Git Bash for Windows, type the following in the terminal:
bash.escapeChar=\
(Note: There is no need to include this in your .bashrc file.)
# GitHub Codespaces: How to store secrets
Instruction on how to store secrets that will be available in GitHub Codespaces. See Managing your account-specific secrets for GitHub Codespaces - GitHub Docs.
# GitHub Codespaces: Running pgadmin in Docker
With the default instructions, running pgadmin in Docker may result in a blank screen after logging into the pgadmin console. To resolve this, add the following two environment variables to your pgadmin configuration to allow it to work with Codespaces’ reverse proxy:
PGADMIN_CONFIG_PROXY_X_HOST_COUNT: 1
PGADMIN_CONFIG_PROXY_X_PREFIX_COUNT: 1
# Docker: Cannot connect to Docker daemon at unix:///var/run/docker.sock. Is the Docker daemon running?
Make sure you're able to start the Docker daemon. Check the issue immediately as described below:
Ensure the Docker daemon is running.
Update WSL in PowerShell with the following command:
wsl --update
# Docker - error during connect: In the default daemon configuration on Windows, the docker client must be run with elevated privileges
If you get this error:
docker: error during connect: In the default daemon configuration on Windows, the docker client must be run with elevated privileges to connect.: Post "http://%2F%2F.%2Fpipe%2Fdocker_engine/v1.24/containers/create": open //./pipe/docker_engine: The system cannot find the file specified.
See 'docker run --help'.
To resolve it on Windows, follow these guidelines based on your version:
Windows 10 Pro / 11 Pro Users:
- Ensure Hyper-V is enabled, as Docker can use it as a backend.
- Follow the Enable Hyper-V Option on Windows 10 / 11 tutorial.
Windows 10 Home / 11 Home Users:
- The 'Home' version doesn't support Hyper-V, so use WSL2 (Windows Subsystem for Linux).
- Refer to install WSL on Windows 11 for detailed instructions.
If you encounter the "WslRegisterDistribution failed with error: 0x800701bc" error:
- Update the WSL2 Linux Kernel by following the guidelines at GitHub: WSL Issue 5393.
# Docker: docker pull dbpage
Whenever a docker pull is performed (either manually or by docker-compose up), it attempts to fetch the given image name from a repository. If the repository is public, the fetch and download occur without any issues.
For instance:
docker pull postgres:13
docker pull dpage/pgadmin4
Be Advised: The Docker images we'll be using throughout the Data Engineering Zoomcamp are all public, unless otherwise specified. This means you are not required to perform a docker login to fetch them.
If you encounter the message:
docker login': denied: requested access to the resource is denied.
This is likely due to a typo in your image name. For instance:
$ docker pull dbpage/pgadmin4
This command will throw an exception:
Error response from daemon: pull access denied for dbpage/pgadmin4, repository does not exist or may require 'docker login': denied: requested access to the resource is denied
This occurs because the actual image name is dpage/pgadmin4, not dbpage/pgadmin4.
How to fix it:
$ docker pull dpage/pgadmin4
Extra Notes: In some professional environments, the Docker image may be in a private repository that your DockerHub username has access to. In this case, you must:
- Execute:
$ docker login - Enter your username and password.
- Then perform the
docker pullagainst that private repository.
# Docker: "permission denied" error when creating a PostgreSQL Docker with a mounted volume on macOS M1
When attempting to run a Docker command similar to the one below:
docker run -it \
-e POSTGRES_USER="root" \
-e POSTGRES_PASSWORD="root" \
-e POSTGRES_DB="ny_taxi" \
-v $(pwd)/ny_taxi_postgres_data:/var/lib/postgresql/data \
-p 5432:5432 \
postgres:13
You encounter the error message:
docker: Error response from daemon: error while creating mount source path '/path/to/ny_taxi_postgres_data': chown /path/to/ny_taxi_postgres_data: permission denied.
Solution
Stop Rancher Desktop:
If you are using Rancher Desktop and face this issue, stop Rancher Desktop to resolve compatibility problems.Install Docker Desktop:
Install Docker Desktop, ensuring that it is properly configured and has the required permissions.Retry Docker Command:
Run the Docker command again after switching to Docker Desktop. This step resolves compatibility issues on some systems.
Note: The issue occurred because Rancher Desktop was in use. Switching to Docker Desktop resolves compatibility problems and allows for the successful creation of PostgreSQL containers with mounted volumes for the New York Taxi Database on macOS M1.
# Docker: can’t delete local folder that mounted to docker volume
When a PostgreSQL Docker container is created, it may create a folder on the local machine to mount to a volume inside the container. This folder is often owned by user 999 and has read and write protection, preventing deletion by conventional means such as dragging it to the trash.
If you encounter an access error or need to delete the folder, you can use the following command:
sudo rm -r -f docker_test/
rm: Command to remove files or directories.-r: Recursively remove directories and their contents.-f: Forcefully remove files/directories without prompting.docker_test/: The folder to be deleted.
# Docker: Docker won't start or is stuck in settings (Windows 10 / 11)
Ensure you are running the latest version of Docker for Windows. Download the updated version from Docker's official site. If the upgrade option in the menu doesn't work, uninstall and reinstall with the latest version.
If Docker is stuck on starting, try switching the containers by right-clicking the docker symbol from the running programs, and switch the containers from Windows to Linux or vice versa.
For Windows 10 / 11 Pro Edition:
# Docker: Should I run docker commands from the windows file system or a file system of a Linux distribution in WSL?
If you're running a Home Edition, you can still make it work with WSL2 (Windows Subsystem for Linux) by following the tutorial here.
If even after making sure your WSL2 (or Hyper-V) is set up accordingly, Docker remains stuck, you can try the following options:
- Reset to Factory Defaults
- Perform a fresh install.
# Docker: The input device is not a TTY (Docker run for Windows)
You may encounter this error:
$ docker run -it ubuntu bash
the input device is not a TTY. If you are using mintty, try prefixing the command with 'winpty'
Solution:
Use
winptybefore the Docker command:$ winpty docker run -it ubuntu bashAlternatively, create an alias:
echo "alias docker='winpty docker'" >> ~/.bashrcor
echo "alias docker='winpty docker'" >> ~/.bash_profile
Source: Stack Overflow
# Docker: Cannot pip install on Docker container (Windows)
You may encounter this error:
Retrying (Retry(total=4, connect=None, read=None, redirect=None, status=None)) after connection broken by 'NewConnectionError('<pip._vendor.urllib3.connection.HTTPSConnection object at 0x7efe331cf790>: Failed to establish a new connection: [Errno -3] Temporary failure in name resolution')': /simple/pandas/
Possible solution:
Run the following command:
winpty docker run -it --dns=8.8.8.8 --entrypoint=bash python:3.9
# Docker: ny_taxi_postgres_data is empty
Even after properly running the Docker script, the folder may appear empty in VS Code. For Windows, try the following steps:
Solution 1:
Run the Docker command with the absolute path quoted in the -v parameter:
winpty docker run -it \
-e POSTGRES_USER="root" \
-e POSTGRES_PASSWORD="root" \
-e POSTGRES_DB="ny_taxi" \
-v "C:\Users\abhin\dataengg\DE_Project_git_connected\DE_OLD\week1_set_up\docker_sql/ny_taxi_postgres_data:/var/lib/postgresql/data" \
-p 5432:5432 \
postgres:13
This should resolve the visibility issue in the VS Code ny_taxi folder.
Note: Ensure the correct direction for the slashes: / versus \.
Solution 2:
Another possible solution for Windows is to finish the folder path with a forward slash /:
docker run -it \
-e POSTGRES_USER="root" \
-e POSTGRES_PASSWORD="root" \
-e POSTGRES_DB="ny_taxi" \
-v /"$(pwd)"/ny_taxi_postgres_data/:/var/lib/postgresql/data/ \
-p 5432:5432 \
postgres:13
These steps should help resolve the issue of the ny_taxi_postgres_data folder appearing empty in your Docker setup.
# Docker: Setting up Docker on Mac
For setting up Docker on macOS, you have two main options:
Download from Docker Website:
- Visit the official Docker website and download the Docker Desktop for Mac as a
.dmgfile. This method is generally reliable and avoids issues related to licensing changes.
- Visit the official Docker website and download the Docker Desktop for Mac as a
Using Homebrew:
Be aware that there can be conflicts when installing with Homebrew, especially between Docker Desktop and command-line tools. To avoid issues:
- Install Docker Desktop first.
- Then install the command line tools.
Commands:
brew install --cask dockerbrew install docker docker-composeFor more detailed issues related to
brew install, refer to this Issue.
For more details, you can check the article on Setting up Docker in macOS.
# Docker - Could not change permissions of directory "/var/lib/postgresql/data": Operation not permitted
docker run -it \
-e POSTGRES_USER="root" \
-e POSTGRES_PASSWORD="admin" \
-e POSTGRES_DB="ny_taxi" \
-v "/mnt/path/to/ny_taxi_postgres_data":"/var/lib/postgresql/data" \
-p 5432:5432 \
postgres:13
Error Message:
The files belonging to this database system will be owned by user "postgres".
The database cluster will be initialized with locale "en_US.utf8".
The default database encoding has accordingly been set to "UTF8".
Data page checksums are disabled.
fixing permissions on existing directory /var/lib/postgresql/data ...
initdb: error: could not change permissions of directory "/var/lib/postgresql/data": Operation not permitted
Solution:
Create a local Docker volume and map it to the Postgres data directory
/var/lib/postgresql/data.- The volume name
dtc_postgres_volume_localmust match in both commands below:
docker volume create --name dtc_postgres_volume_local -d local- The volume name
Run the Docker container using the created volume:
docker run -it \ -e POSTGRES_USER="root" \ -e POSTGRES_PASSWORD="root" \ -e POSTGRES_DB="ny_taxi" \ -v dtc_postgres_volume_local:/var/lib/postgresql/data \ -p 5432:5432 \ postgres:13Verify the command works in Docker Desktop under Volumes. The
dtc_postgres_volume_localshould be listed, but the folderny_taxi_postgres_datawill be empty as an alternative configuration is used.
Alternate Error:
initdb: error: directory "/var/lib/postgresql/data" exists but is not empty
To resolve this, either remove or empty the directory "/var/lib/postgresql/data", or run initdb.
# Docker: invalid reference format: repository name must be lowercase (Mounting volumes with Docker on Windows)
Mapping volumes on Windows can be tricky. If the approach shown in the course video doesn't work for you, consider the following suggestions:
Move your data to a directory without spaces. For example, move from
C:/Users/Alexey Grigorev/git/...toC:/git/....Replace the
-vpart with one of these options:-v /c:/some/path/ny_taxi_postgres_data:/var/lib/postgresql/data -v //c:/some/path/ny_taxi_postgres_data:/var/lib/postgresql/data -v /c/some/path/ny_taxi_postgres_data:/var/lib/postgresql/data -v //c/some/path/ny_taxi_postgres_data:/var/lib/postgresql/data --volume //driveletter/path/ny_taxi_postgres_data/:/var/lib/postgresql/dataAdd
winptybefore the whole command:winpty docker run -it \ -e POSTGRES_USER="root" \ -e POSTGRES_PASSWORD="root" \ -e POSTGRES_DB="ny_taxi" \ -v /c:/some/path/ny_taxi_postgres_data:/var/lib/postgresql/data \ -p 5432:5432 \ postgres:1Try adding quotes:
-v "/c:/some/path/ny_taxi_postgres_data:/var/lib/postgresql/data" -v "//c:/some/path/ny_taxi_postgres_data:/var/lib/postgresql/data" -v “/c/some/path/ny_taxi_postgres_data:/var/lib/postgresql/data" -v "//c/some/path/ny_taxi_postgres_data:/var/lib/postgresql/data" -v "c:\some\path\ny_taxi_postgres_data":/var/lib/postgresql/dataNote: If Windows automatically creates a folder called
ny_taxi_postgres_data;C, it suggests a problem with volume mapping. Try deleting both folders and replacing the-vpart with other options. Using//c/instead of/c/might work, as it creates the correct folderny_taxi_postgres_data.A possible solution is using
"$(pwd)"/ny_taxi_postgres_data:/var/lib/postgresql/dataand pay attention to the placement of quotes.If none of these work, use a volume name instead of the path:
-v ny_taxi_postgres_data:/var/lib/postgresql/dataFor Mac, you can wrap
$(pwd)with quotes:docker run -it \ -e POSTGRES_USER="root" \ -e POSTGRES_PASSWORD="root" \ -e POSTGRES_DB="ny_taxi" \ -v "$(pwd)"/ny_taxi_postgres_data:/var/lib/postgresql/data \ -p 5432:5432 \ postgres:13
Source: StackOverflow
# Docker: Error response from daemon: invalid mode: \Program Files\Git\var\lib\postgresql\data.
Change the mounting path. Replace it with one of the following:
-v /e/zoomcamp/...:/var/lib/postgresql/data
Or
-v /c:/.../ny_taxi_postgres_data:/var/lib/postgresql/data
(Note: Include a leading slash in front of c:)
# Docker: Error response from daemon: error while creating buildmount source
You may get this error:
error while creating buildmount source path '/run/desktop/mnt/host/c/<your path>': mkdir /run/desktop/mnt/host/c: file exists
When you encounter the error above while rerunning your Docker command, it indicates that you should not mount on the second run. Here’s the initial problematic command:
docker run -it \
-e POSTGRES_USER="root" \
-e POSTGRES_PASSWORD="root" \
-e POSTGRES_DB="ny_taxi" \
-v <your path>:/var/lib/postgresql/data \
-p 5432:5432 \
postgres:13
To resolve the issue, use the revised command without the volume mount:
docker run -it \
-e POSTGRES_USER="root" \
-e POSTGRES_PASSWORD="root" \
-e POSTGRES_DB="ny_taxi" \
-p 5432:5432 \
postgres:13
# Docker: build error: error checking context: 'can't stat '/home/user/repos/data-engineering/week_1_basics_n_setup/2_docker_sql/ny_taxi_postgres_data''.
This error appeared when running the command:
docker build -t taxi_ingest:v001 .
The issue often arises because the user ID of the directory ny_taxi_postgres_data was changed, causing permission errors when accessing it. To resolve this error, use a directory containing only the necessary files, Dockerfile and ingest_data.py.
If you need to change permissions, use the following command on Ubuntu:
sudo chown -R $USER dir_path
On Windows, follow the instructions in this guide: The Geek Page.
For more information, refer to this explanation on Stack Overflow: Docker build error checking context.
# Docker: ERRO[0000] error waiting for container: context canceled
You might have installed Docker via snap. Run the following command to verify:
sudo snap status docker
If you receive the response:
error: unknown command "status", see 'snap help'.
Then uninstall Docker and install it via the official website.
Error message: "Bind for 0.0.0.0:5432 failed: port is already allocated."
# Docker: build error checking context: can’t stat ‘/home/fhrzn/Projects/…./ny_taxi_postgres_data’
This issue occurs due to insufficient authorization rights to the host folder, which may cause it to appear empty.
Solution:
Add permission for everyone to the folder:
sudo chmod -R 777 <path_to_folder>
Example:
sudo chmod -R 777 ny_taxi_postgres_data/
# Docker: failed to solve with frontend dockerfile.v0: failed to read dockerfile: error from sender: open ny_taxi_postgres_data: permission denied.
This issue occurs on Ubuntu/Linux systems when attempting to rebuild the Docker container.
$ docker build -t taxi_ingest:v001 .
A folder is created to host the Docker files. When the build command is executed again, a permission error may occur because there are no permissions on this new folder. To resolve this, grant permissions by running the command:
$ sudo chmod -R 755 ny_taxi_postgres_data
If issues persist, use:
$ sudo chmod -R 777 ny_taxi_postgres_data
Note: 755 grants write access only to the owner.
# Docker: Docker network name
Get the network name via:
docker network ls
For more details, refer to the Docker network ls documentation.
# Docker: Error response from daemon: Conflict. The container name "pg-database" is already in use by container "xxx". You have to remove (or rename) that container to be able to reuse that name.
Sometimes, when you try to restart a Docker container configured with a network name, the error message appears.
To resolve this issue:
If the container is in a running state, stop it using:
docker stop <container_name>Then remove the container:
docker rm pg-database
Alternatively, you can use docker start instead of docker run to restart the Docker container without removing it.
# Docker: ingestion when using docker-compose could not translate host name
Typical error:
n.exc.OperationalError: (psycopg2.OperationalError) could not translate host name "pgdatabase" to address: Name or service not known
Solution:
- Run
docker-compose up -dto start your containers. - Check which network is created by Docker, as it may differ from your expectations.
- Use the actual network name in your ingestion script instead of "pg-network".
- Confirm the correct database service name, replacing "pgdatabase" accordingly.
Example:
- "pg-network" might become "2docker_default".
# Docker: Cannot install docker on MacOS/Windows 11 VM running on top of Linux (due to Nested virtualization).
Before starting your VM, you need to enable nested virtualization. Run the following commands based on your CPU:
For Intel CPU:
modprobe -r kvm_intel modprobe kvm_intel nested=1For AMD CPU:
modprobe -r kvm_amd modprobe kvm_amd nested=1
# Docker: Connecting from VS Code
It’s very easy to manage your Docker container, images, network, and compose projects from VS Code.
Install the official extension and launch it from the left side icon.
It will work even if your Docker runs on WSL2, as VS Code can easily connect with your Linux.
# Docker: How to stop a container?
Use the following command:
docker stop <container_id>
# Docker: PostgreSQL Database directory appears to contain a database. Database system is shut down
When you see this in logs, your container with PostgreSQL is not accepting any requests. Attempting to connect may result in the error:
connection failed: server closed the connection unexpectedly
This probably means the server terminated abnormally before or while processing the request.
To resolve this issue:
Delete Data Directory: Delete the directory with data (the one you map to the container using the
-vflag) and restart the container.Preserve Critical Data: If your data is critical, you may be able to reset the write-ahead log from within the Docker container. For more details, see here.
docker run -it \ -e POSTGRES_USER="root" \ -e POSTGRES_PASSWORD="root" \ -e POSTGRES_DB="ny_taxi" \ -v $(pwd)/ny_taxi_postgres_data:/var/lib/postgresql/data \ -p 5432:5432 \ --network pg-network \ postgres:13 \ /bin/bash -c 'gosu postgres pg_resetwal /var/lib/postgresql/data'
# Docker: Docker not installable on Ubuntu
On some versions of Ubuntu, the snap command can be used to install Docker.
sudo snap install docker
# Docker-Compose: mounting error
error: could not change permissions of directory "/var/lib/postgresql/data": Operation not permitted
If you have used the previous answer and created a local Docker volume, then you need to inform the compose file about the named volume:
dtc_postgres_volume_local: # Define the named volume here
- Services mentioned in the compose file automatically become part of the same network.
Steps:
Use the command:
docker volume inspect dtc_postgres_volume_localto see the location by checking the value of
Mountpoint.In some cases, after running
docker compose up, the mounting directory created is nameddocker_sql_dtc_postgres_volume_localinstead of the existingdtc_postgres_volume_local.Rename the existing
dtc_postgres_volume_localtodocker_sql_dtc_postgres_volume_local:- Be careful when performing this operation.
Remove the newly created one.
Run
docker compose upagain and check if the table is there.
# Docker-Compose: Error translating host name to address
Couldn’t translate host name to address
# Docker-Compose: Data retention (could not translate host name "pg-database" to address: Name or service not known)
Make sure the PostgreSQL database is running. Use the command to start containers in detached mode:
docker-compose up -d
Example output:
% docker compose up -d
[+] Running 2/2
⠿ Container pg-admin Started
⠿ Container pg-database Started
To view the containers use:
docker ps
Example output:
% docker ps
CONTAINER ID IMAGE COMMAND CREATED STATUS PORTS NAMES
faf05090972e postgres:13 "docker-entrypoint.s…" 39 seconds ago Up 37 seconds 0.0.0.0:5432->5432/tcp pg-database
6344dcecd58f dpage/pgadmin4 "/entrypoint.sh" 39 seconds ago Up 37 seconds 443/tcp, 0.0.0.0:8080->80/tcp pg-adminhw
To view logs for a container:
docker logs <containerid>
Example logs for PostgreSQL:
% docker logs faf05090972e
PostgreSQL Database directory appears to contain a database; Skipping initialization
2022-01-25 05:58:45.948 UTC [1] LOG: starting PostgreSQL 13.5 (Debian 13.5-1.pgdg110+1) on aarch64-unknown-linux-gnu, compiled by gcc (Debian 10.2.1-6) 10.2.1 20210110, 64-bit
2022-01-25 05:58:45.948 UTC [1] LOG: listening on IPv4 address "0.0.0.0", port 5432
2022-01-25 05:58:45.948 UTC [1] LOG: listening on IPv6 address "::", port 5432
2022-01-25 05:58:45.954 UTC [1] LOG: listening on Unix socket "/var/run/postgresql/.s.PGSQL.5432"
2022-01-25 05:58:45.984 UTC [28] LOG: database system was interrupted; last known up at 2022-01-24 17:48:35 UTC
2022-01-25 05:58:48.581 UTC [28] LOG: database system was not properly shut down; automatic recovery in progress
2022-01-25 05:58:48.602 UTC [28] LOG: redo starts at 0/872A5910
2022-01-25 05:59:33.726 UTC [28] LOG: invalid record length at 0/98A3C160: wanted 24, got 0
2022-01-25 05:59:33.726 UTC [28] LOG: redo done at 0/98A3C128
2022-01-25 05:59:48.051 UTC [1] LOG: database system is ready to accept connections
If docker ps doesn’t show pg-database running, use:
docker ps -a
- This will show all containers, either running or stopped.
- Get the container ID for
pg-database-1and run the appropriate command.
If you lose database data after executing docker-compose up and cannot successfully execute your ingestion script due to the following error:
sqlalchemy.exc.OperationalError: (psycopg2.OperationalError) could not translate host name /data_pgadmin:/var/lib/pgadmin"pg-database" to address: Name or service not known
- Docker Compose may be creating its own default network since it is no longer specified in the command or file.
- Check logs after executing
docker-compose upto find the network name and change the network name argument in your ingestion script.
If problems persist with pgcli, consider using HeidiSQL.
# Docker-Compose: Hostname does not resolve
When encountering the error:
Error response from daemon: network 66ae65944d643fdebbc89bd0329f1409dec2c9e12248052f5f4c4be7d1bdc6a3 not found
Try the following steps:
- Run
docker ps -ato see all stopped and running containers. - Remove all containers to clean up the environment.
- Execute
docker-compose up -dagain.
If facing issues connecting to the server at localhost:8080 with the error:
Unable to connect to server: could not translate host name 'pg-database' to address: Name does not resolve
Consider these solutions:
- Use a new hostname without dashes, e.g.,
pgdatabase. - Make sure to specify the Docker network and use the same network in both containers in your
docker-compose.ymlfile.
Example docker-compose.yml:
services:
pgdatabase:
image: postgres:13
environment:
- POSTGRES_USER=root
- POSTGRES_PASSWORD=root
- POSTGRES_DB=ny_taxi
volumes:
- "./ny_taxi_postgres_data:/var/lib/postgresql/data:rw"
ports:
- "5431:5432"
networks:
- pg-network
pgadmin:
image: dpage/pgadmin4
environment:
- PGADMIN_DEFAULT_EMAIL=admin@admin.com
- PGADMIN_DEFAULT_PASSWORD=root
ports:
- "8080:80"
networks:
- pg-network
networks:
pg-network:
name: pg-network
# Docker-Compose: PgAdmin – no database in PgAdmin
When you log into PgAdmin and see an empty database, the following solution can help:
Run:
docker-compose up
And at the same time run:
docker build -t taxi_ingest:v001 .
# NETWORK NAME IS THE SAME AS THAT CREATED BY DOCKER COMPOSE
docker run -it \
--network=pg-network \
taxi_ingest:v001 \
--user=postgres \
--password=postgres \
--host=db \
--port=5432 \
--db=ny_taxi \
--table_name=green_tripdata \
--url=${URL}
It's important to use the same --network as stated in the docker-compose.yaml file.
The docker-compose.yaml file might not specify a network, as shown below:
services:
db:
container_name: postgres
image: postgres:17-alpine
environment:
...
ports:
- '5433:5432'
volumes:
- ...
pgadmin:
container_name: pgadmin
image: dpage/pgadmin4:latest
environment:
...
ports:
- '8080:80'
volumes:
- ...
volumes:
vol-pgdata:
name: vol-pgdata
vol-pgadmin_data:
name: vol-pgadmin_data
If the network name is not specified, it is generated automatically: The name of the directory containing the docker-compose.yaml file in lowercase + _default.
You can find the network’s name when running docker-compose up:
pg-database Pulling pg-database Pulled
Network week_1_default Creating
Network week_1_default Created
# Docker-Compose: Persist PGAdmin docker contents on GCP
One common issue when running Docker Compose on GCP is that PostgreSQL might not persist its data to the specified path. For example:
services:
...
pgadmin:
...
volumes:
- "./pgadmin:/var/lib/pgadmin:wr"
This setup might not work. To resolve this, use Docker Volume to make the data persist:
services:
...
pgadmin:
...
volumes:
- pgadmin:/var/lib/pgadmin
volumes:
pgadmin:
This configuration change ensures the persistence of the PGAdmin data on GCP.
# Docker: Docker engine stopped_failed to fetch extensions
The Docker engine may crash continuously and fail to work after restart. You might see error messages like "docker engine stopped" and "failed to fetch extensions" repeatedly on the screen.
Solution:
- Check if you have the latest version of Docker installed. Update Docker if necessary.
- If the problem persists, consider reinstalling Docker.
- Note: You will need to fetch images again, but there should be no other issues.
# Docker-Compose: Persist PGAdmin configuration
To persist pgAdmin configuration, such as the server name, modify your docker-compose.yml by adding a "volumes" section:
services:
pgdatabase:
[...]
pgadmin:
image: dpage/pgadmin4
environment:
- PGADMIN_DEFAULT_EMAIL=admin@admin.com
- PGADMIN_DEFAULT_PASSWORD=root
volumes:
- "./pgAdmin_data:/var/lib/pgadmin/sessions:rw"
ports:
- "8080:80"
In the example above, "pgAdmin_data" is a folder on the host machine, and "/var/lib/pgadmin/sessions" is the session settings folder in the pgAdmin container.
Before running docker-compose up on the YAML file, provide the pgAdmin container with access permissions to the "pgAdmin_data" folder. The container runs with a username "5050" and user group "5050". Use the following command to set permissions:
sudo chown -R 5050:5050 pgAdmin_data
# Docker-Compose - dial unix /var/run/docker.sock: connect: permission denied
This happens if you did not create the docker group and add your user. Follow these steps from the link: guides/docker-without-sudo.md at main · sindresorhus/guides · GitHub
- Press
Ctrl+Dto log out and log back in again.
If you are tired of having to set up your database connection each time you start the containers, create a volume for pgAdmin:
In your docker-compose.yaml file, add the following under your pgAdmin service:
services:
pgadmin:
volumes:
- type: volume
source: pgadmin_data
target: /var/lib/pgadmin
Also, add the following to the end of the file:
volumes:
pgadmin_data:
This configuration will maintain the state so that pgAdmin remembers your previous connections.
# Docker: docker-compose still not available after changing .bashrc
This issue can occur after installing Docker Compose in a Google Cloud VM, as demonstrated in video 1.4.1.
If the downloaded Docker Compose file from GitHub is named docker-compose-linux-x86_64, you may need to rename it for convenience. Here's how to resolve the issue:
Rename
docker-compose-linux-x86_64todocker-composeusing the following command:mv docker-compose-linux-x86_64 docker-compose
By doing this, you can use the docker-compose command directly.
# Docker-Compose: Error getting credentials after running docker-compose up -d
Installing pass via sudo apt install pass helped to solve the issue. More about this can be found here: https://github.com/moby/buildkit/issues/1078
# Docker-Compose: Errors pertaining to docker-compose.yml and pgadmin setup
For those experiencing problems with Docker Compose, getting data in PostgreSQL, and similar issues, follow these steps:
- Create a new volume on Docker, either using the command line or Docker Desktop app.
- Modify your
docker-compose.ymlfile as needed to fix any setup issues. - Set
low_memory=Falsewhen importing the CSV file using pandas:
df = pd.read_csv('yellow_tripdata_2021-01.csv', nrows=1000, low_memory=False)
- Use the specified function in your
upload-data.ipynbfor better tracking of the ingestion process.
from time import time
counter = 0
time_counter = 0
while True:
t_start = time()
df = next(df_iter)
df.tpep_pickup_datetime = pd.to_datetime(df.tpep_pickup_datetime)
df.tpep_dropoff_datetime = pd.to_datetime(df.tpep_dropoff_datetime)
df.to_sql(name='yellow_taxi_data', con=engine, if_exists='append')
t_end = time()
t_elapsed = t_end - t_start
print('Chunk Insertion Done! Time taken: %.2f seconds' %(t_elapsed))
counter += 1
time_counter += t_elapsed
if counter == 14:
print('All Chunks Inserted! Total Time Taken: %.2f seconds' %(time_counter))
break
Order of Execution:
- Open the terminal in the
2_docker_sqlfolder and run:docker compose up - Ensure no other containers are running except the ones you just executed (pgAdmin and pgdatabase).
- Open Jupyter Notebook and begin the data ingestion.
- Open pgAdmin and set up a server. Make sure you use the same configurations as your
docker-compose.ymlfile, such as the same name (pgdatabase), port, and database name (ny_taxi).
# Docker: Compose up -d error getting credentials - err: exec: "docker-credential-desktop": executable file not found in %PATH%, out: ``
To resolve this error, follow these steps:
- Locate the
config.jsonfile for Docker, typically found in your home directory atUsers/username/.docker. - Modify the
credsStoresetting tocredStore. - Save the file and re-run your Docker Compose command.
# Docker-Compose: Which docker-compose binary to use for WSL?
To determine which docker-compose binary to download from Docker Compose releases, you can check your system with the following commands:
To check the system type:
uname -s # This will most likely return 'Linux'To check the system architecture:
uname -m # This will return your system's 'flavor'
Alternatively, you can use the following command to download docker-compose directly:
sudo curl -L "https://github.com/docker/compose/releases/download/1.29.2/docker-compose-$(uname -s)-$(uname -m)" -o /usr/local/bin/docker-compose
# Docker-Compose - Error undefined volume in Windows/WSL
If you wrote the docker-compose.yaml file exactly like the video, you might run into an error:
service "pgdatabase" refers to undefined volume dtc_postgres_volume_local: invalid compose project
To resolve this, include the volume definition in your docker-compose.yaml file by adding:
dt_postgres_volume_local:
This should be added under the volumes section. Make sure your file looks similar to this:
volumes:
dtc_postgres_volume_local:
# Docker-Compose: cannot execute binary file: Exec format error
This error indicates that the docker-compose executable cannot be opened in the current OS. Ensure that the file you download from GitHub matches your system environment.
As of 2025/1/17, docker-compose (v2.32.4) docker-compose-linux-aarch64 does not work. Try v2.32.3 docker-compose-linux-x86_64.
# Docker: Postgres container fails to launch with exit code (1) when attempting to compose
This issue arises because the Postgres database is not initialized before executing docker-compose up -d. While there are other potential solutions discussed in this thread, you can resolve it by initializing the database first. Then, the Docker Compose will work as expected.
docker run -it \
-e POSTGRES_USER="root" \
-e POSTGRES_PASSWORD="root" \
-e POSTGRES_DB="ny_taxi" \
-v $(pwd)/ny_taxi_data:/var/lib/postgresql/data \
-p 5432:5432 \
--network=pg-network \
--name=pg_database \
postgres:13
# WSL: Docker directory permissions error
initdb: error: could not change permissions of directory
WSL and Windows do not manage permissions in the same way, causing conflict if using the Windows file system rather than the WSL file system.
Solution: Use Docker volumes.
Volume is used for storage of persistent data and not for transferring files. A local volume is unnecessary.
This resolves permission issues and allows for better management of volumes.
Note: The user: is not necessary if using Docker volumes but is required if using a local drive.
services:
postgres:
image: postgres:15-alpine
container_name: postgres
user: "0:0"
environment:
- POSTGRES_USER=postgres
- POSTGRES_PASSWORD=postgres
- POSTGRES_DB=ny_taxi
volumes:
- "pg-data:/var/lib/postgresql/data"
ports:
- "5432:5432"
networks:
- pg-network
pgadmin:
image: dpage/pgadmin4
container_name: pgadmin
user: "${UID}:${GID}"
environment:
- PGADMIN_DEFAULT_EMAIL=email@some-site.com
- PGADMIN_DEFAULT_PASSWORD=pgadmin
volumes:
- "pg-admin:/var/lib/pgadmin"
ports:
- "8080:80"
networks:
- pg-network
networks:
pg-network:
name: pg-network
volumes:
pg-data:
pg-admin:
# WSL: Insufficient system resources exist to complete the requested service.
Cause:
This error occurs because some applications are not updated. Specifically, check for any pending updates for Windows Terminal, WSL, and Windows Security updates.
Solution:
To update Windows Terminal:
- Open the Microsoft Store.
- Go to your library of installed apps.
- Search for Windows Terminal.
- Update the app.
- Restart your system to apply the changes.
For updating Windows Security updates:
- Go to Windows Updates settings.
- Check for any pending updates, especially security updates.
- Restart your system once the updates are downloaded and installed successfully.
# WSL: WSL integration with distro Ubuntu unexpectedly stopped with exit code 1.
Upon restarting, the same issue appears and occurs unexpectedly on Windows.
Solutions:
Fixing DNS Issue
This solution is credited to reddit and has worked for some users.
reg add "HKLM\System\CurrentControlSet\Services\Dnscache" /v "Start" /t REG_DWORD /d "4" /fRestart your computer and then re-enable it with the following command:
reg add "HKLM\System\CurrentControlSet\Services\Dnscache" /v "Start" /t REG_DWORD /d "2" /fRestart your OS again. It should work.
Switch to Linux Containers
- Right-click on the running Docker icon (next to the clock).
- Choose "Switch to Linux containers."
bash: conda: command not found
Database is uninitialized and superuser password is not specified.
Database is uninitialized and superuser password is not specified.
# WSL: Permissions too open at Windows
Issue when trying to run the GPC VM through SSH via WSL2, likely because WSL2 isn’t looking for .ssh keys in the correct folder. The command attempted:
ssh -i gpc [username]@[my external IP]
Solutions
Use
sudoCommandTry using
sudobefore executing the command:sudo ssh -i gpc [username]@[my external IP]Change Permissions
Navigate to your folder and change the permissions for the private key SSH file:
chmod 600 gpcCreate a
.sshFolder in WSL2Navigate to your home directory:
cd ~Create a
.sshfolder:mkdir .sshCopy the content from the Windows
.sshfolder to the newly created.sshfolder:cp -r /mnt/c/Users/YourUsername/.ssh/* ~/.ssh/Adjust the permissions of the files and folders in the
.sshdirectory if necessary.
# WSL: Could not resolve host name
WSL2 may not be referencing the correct .ssh/config path from Windows. You can create a config file in the home directory of WSL2 by following these steps:
Navigate to your home directory:
cd ~Create the
.sshdirectory:mkdir .sshCreate a
configfile in the.sshfolder with the following content:HostName [GPC VM external IP] User [username] IdentityFile ~/.ssh/[private key]
# PGCLI - connection failed: :1), port 5432 failed: could not receive data from server: Connection refused could not send SSL negotiation packet: Connection refused
To resolve the connection failure with PGCLI, use the following command to connect via socket:
pgcli -h 127.0.0.1 -p 5432 -u root -d ny_taxi
Ensure the database server is running and properly configured to accept connections.
# PGCLI: Should we run pgcli inside another docker container?
In this section of the course, the 5432 port of PostgreSQL is mapped to your computer’s 5432 port. This means you can access the PostgreSQL database via pgcli directly from your computer.
So, no, you don’t need to run it inside another container. Your local system will suffice.
# PGCLI - FATAL: password authentication failed for user "root" (You already have Postgres)
For a more visual and detailed explanation, feel free to check the video 1.4.2 - Port Mapping and Networks in Docker.
If you want to debug the issue on MacOS, you can try the following steps:
Check if something is blocking your port:
Use the
lsofcommand to find out which application is using a specific port on your local machine:lsof -i :5432List running PostgreSQL services:
Use
launchctlto list running postgres services on your local machine.Unload the running service:
Unload the launch agent for the PostgreSQL service, which will stop the service and free up the port:
launchctl unload -w ~/Library/LaunchAgents/homebrew.mxcl.postgresql.plistRestart the service:
launchctl load -w ~/Library/LaunchAgents/homebrew.mxcl.postgresql.plistChange the port:
Changing the port from
5432:5432to5431:5432can help avoid this error.
# PGCLI - PermissionError: [Errno 13] Permission denied: '/some/path/.config/pgcli'
I encountered this error:
pgcli -h localhost -p 5432 -U root -d ny_taxi
Traceback (most recent call last):
File "/opt/anaconda3/bin/pgcli", line 8, in <module>
sys.exit(cli())
File "/opt/anaconda3/lib/python3.9/site-packages/click/core.py", line 1128, in __call__
return self.main(*args, **kwargs)
File "/opt/anaconda3/lib/python3.9/site-packages/click/core.py", line 1053, in main
rv = self.invoke(ctx)
File "/opt/anaconda3/lib/python3.9/site-packages/click/core.py", line 1395, in invoke
return ctx.invoke(self.callback, **ctx.params)
File "/opt/anaconda3/lib/python3.9/site-packages/click/core.py", line 754, in invoke
return __callback(*args, **kwargs)
File "/opt/anaconda3/lib/python3.9/site-packages/pgcli/main.py", line 880, in cli
os.makedirs(config_dir)
File "/opt/anaconda3/lib/python3.9/os.py", line 225, in makedirspython
mkdir(name, mode)
PermissionError: [Errno 13] Permission denied: '/Users/vray/.config/pgcli'
Solution 1:
This error indicates that your user doesn’t have the necessary permissions to access or modify the directory or file (/some/path/.config/pgcli). This can occur in Docker environments when privileges are assigned to root instead of the current user.
To resolve this:
Check the file permissions:
ls -l /some/path/.config/pgcliChange the ownership/permissions so that your user has the necessary permissions:
sudo chown -R user_name /Users/user_name/.configsudostands for Super User DO.chownmeans change owner.-Rapplies recursively.user_nameis your PC username (e.g., vray).
Solution 2:
Make sure you install pgcli without using sudo. The recommended approach is to use conda/anaconda to avoid affecting your system Python.
If conda install gets stuck at "Solving environment," try these alternatives:
https://stackoverflow.com/questions/63734508/stuck-at-solving-environment-on-anaconda
# PGCLI - no pq wrapper available.
Error:
ImportError: no pq wrapper available.
Problem Details:
- Could not import
\dt opg 'c' implementation: No module named 'psycopg_c'couldn't import psycopg 'binary' implementation: No module named 'psycopg_binary'couldn't import psycopg 'python' implementation: libpq library not found
Solution:
Check Python Version:
Ensure your Python version is at least 3.9. The
'psycopg2-binary'might fail to install on older versions like 3.7.3.$ python -VEnvironment Setup:
If your Python version is not 3.9, create a new environment:
$ conda create --name de-zoomcamp python=3.9 $ conda activate de-zoomcamp
Install Required Libraries:
Install Postgres libraries:
$ pip install psycopg2-binary $ pip install psycopg_binary
Upgrade pgcli:
If the above steps do not work, try upgrading
pgcli:$ pip install --upgrade pgcli
Install pgcli via Conda:
Make sure to also install
pgcliusing conda:$ conda install -c conda-forge pgcli
If you follow these steps, you should be able to resolve the issue.
# PGCLI - stuck on password prompt
If your Bash prompt is stuck on the password command for postgres:
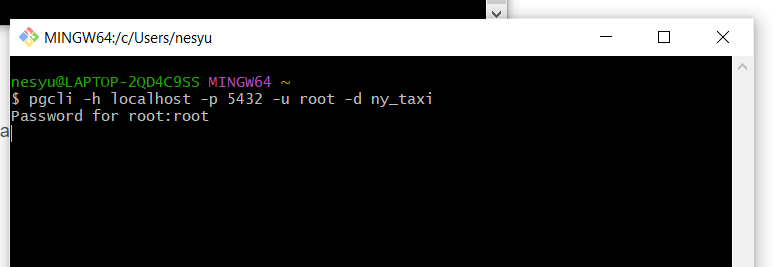
Use winpty:
winpty pgcli -h localhost -p 5432 -u root -d ny_taxi
Alternatively, try using Windows Terminal or the terminal in VS Code.
# PGCLI -connection failed: FATAL: password authentication failed for user "root"
The error above was faced continually despite inputting the correct password.
Stop the PostgreSQL service on Windows
Using WSL: Completely uninstall PostgreSQL 12 from Windows and install
postgresql-clienton WSL:
sudo apt install postgresql-client-common postgresql-client libpq-dev
Change the port of the Docker container
Keep the Database Connection:
If you encounter the error:
PGCLI -connection failed: FATAL: password authentication failed for user "root"
It might be because the connection to the Postgres:13 image was closed. Ensure you keep the database connected in order to continue with the tutorial steps, using the following command:
docker run -it \
-e POSTGRES_USER=root \
-e POSTGRES_PASSWORD=root \
-e POSTGRES_DB=ny_taxi \
-v d:/git/data-engineering-zoomcamp/week_1/docker_sql/ny_taxi_postgres_data:/var/lib/postgresql/data \
-p 5432:5432 \
postgres:13
You should see this
2024-01-26 20:14:43.124 UTC [1] LOG: database system is ready to accept connections
Change the Port for Docker PostgreSQL:
After running the command
pgcli -h localhost -p 5432 -u root -d ny_taxi, if prompted for a password, the error may persist due to local Postgres installation. To resolve this port conflict between host and container:- Configure your Docker PostgreSQL container to use a different port. Map it to a different port on your host machine:
docker run -it \
-e POSTGRES_USER="root" \
-e POSTGRES_PASSWORD="root" \
-e POSTGRES_DB="ny_taxi" \
-v c:/workspace/de-zoomcamp/1_intro_to_data_engineering/docker_sql/ny_taxi_postgres_data:/var/lib/postgresql/data \
-p 5433:5432 \
postgres:13
5433refers to the port on the host machine.5432refers to the port inside the Docker Postgres container.
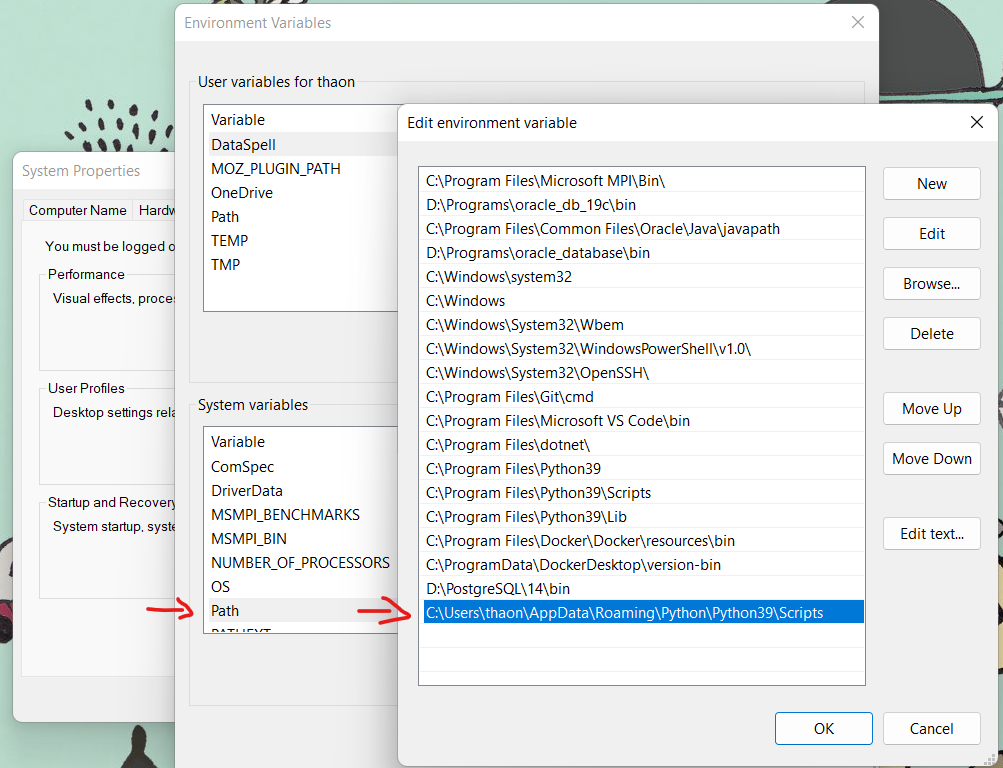
# PGCLI - pgcli: command not found
Problem
If you have already installed pgcli but Bash or the Windows Terminal doesn't recognize the command:
- On Git Bash:
bash: pgcli: command not found - On Windows Terminal:
pgcli: The term 'pgcli' is not recognized…
Solution
Try adding the Python path to the Windows PATH variable:
- Use the command to get the location:
pip list -v - Copy the path, which looks like:
C:\Users\...\AppData\Roaming\Python\Python39\site-packages - Replace
site-packageswithScripts:C:\Users\...\AppData\Roaming\Python\Python39\Scripts
It might be that Python is installed elsewhere. For example, it could be under:
c:\python310\lib\site-packages
In that case, you should add:
c:\python310\lib\Scriptsto PATH.
Instructions
- Add the determined path to
Path(orPATH) in System Variables.
Reference
# PGCLI - running in a Docker container
If running pgcli locally causes issues or you do not want to install it on your machine, you can use it within a Docker container instead.
Below is the usage with values used in the course videos for:
- Network name (Docker network)
- Postgres-related variables for pgcli
- Hostname
- Username
- Port
- Database name
docker run -it --rm --network pg-network ai2ys/dockerized-pgcli:4.0.1
Then execute the following pgcli command:
pgcli -h pg-database -U root -p 5432 -d ny_taxi
You'll be prompted for the password for the user root.
Example Output:
Server: PostgreSQL 16.1 (Debian 16.1-1.pgdg120+1)
Version: 4.0.1
Home: [pgcli.com](http://pgcli.com)
To list tables:
root@pg-database:ny_taxi> \dt
+--------+------------------+-------+-------+
| Schema | Name | Type | Owner |
|--------+------------------+-------+-------|
| public | yellow_taxi_data | table | root |
+--------+------------------+-------+-------+
SELECT 1
Time: 0.009s
root@pg-database:ny_taxi>
# RRPGCLI: Case sensitive use of “Quotations” around columns with capital letters
PULocationID will not be recognized, but "PULocationID" will be. This is because unquoted identifiers are case insensitive. See docs.
# PGCLI - error column c.relhasoids does not exist
When using the command \d <database name> you get the error column c.relhasoids does not exist.
Resolution:
- Uninstall pgcli.
- Reinstall pgcli.
- Restart your PC.
# Postgres: bind: address already in use
Issue
When attempting to start the Docker Postgres container, you may encounter the error message:
Error - postgres port is already in use.
Solutions
Option 1: Identify and Stop the Service
Determine which service is using the port by running:
sudo lsof -i :5432Stop the service that is using the port:
sudo service postgresql stop
Option 2: Map to a Different Port
For a more long-term solution, consider mapping to a different port:
- Map local port 5433 to container port 5432 in your Docker configuration (
Dockerfileordocker-compose.yml). - If using a VM, ensure that port 5433 is forwarded in the host machine configuration.
This approach prevents conflicts and allows the Docker Postgres container to run without interruption.
# PGCLI - After installing PGCLI and checking with `pgcli --help` we get the error: `ImportError: no pq wrapper available`
The error persists because the psycopg library cannot find the required libpq library. Ensure the required PostgreSQL client library is installed:
sudo apt install libpq-dev
Rebuild psycopg:
Uninstall the existing packages:
pip uninstall psycopg psycopg_binary psycopg_c -yReinstall psycopg:
pip install psycopg --no-binary psycopg
The issue should be resolved by now. However, if you still encounter the error:
ModuleNotFoundError: No module named 'psycopg2'
Then run the following:
pip install psycopg2-binary
# Postgres - OperationalError: (psycopg2.OperationalError) connection to server at "localhost" (::1), port 5432 failed: FATAL: password authentication failed for user "root"
This error occurs when uploading data via a connection in Jupyter Notebook:
engine = create_engine('postgresql://root:root@localhost:5432/ny_taxi')
Possible Solutions:
Port Conflict:
- Port 5432 might be occupied by another Postgres installation on your local machine. This can lead to your connection not reaching Docker.
- Try using a different port, such as 5431, or verify the port mapping.
- Alternatively, remove any old or unnecessary Postgres installations if they're not in use.
Windows Service Check:
- Check for any running services on Windows that might be using Postgres.
- Stopping such services might resolve the issue.
# Postgres: connection failed: connection to server at "127.0.0.1", port 5432 failed: FATAL: password authentication failed for user "root"
To resolve the issue of a failed connection to PostgreSQL due to password authentication, consider the following steps:
Check Port Usage: Ensure that port 5432 is properly forwarded. If it is being used by another process, follow these steps to kill it:
sudo lsof -i :5432 sudo kill -9 PIDFor Windows Users: If PostgreSQL is running locally and pgAdmin4 is using the 5432 port, follow these instructions:
- Press Win + R to open the Run dialog.
- Type
services.mscand press Enter. - In the Services window, scroll down to find a service named like
PostgreSQL,postgresql-x64-13, or similar, depending on your PostgreSQL version. - Right-click the PostgreSQL service and select Stop.
# Postgres - OperationalError: (psycopg2.OperationalError) connection to server at "localhost" (::1), port 5432 failed: FATAL: role "root" does not exist
This error can occur in the following scenarios:
- Using
pgcli:pgcli -h localhost -p 5432 -U root -d ny_taxi - Uploading data via a connection in a Jupyter notebook:
engine = create_engine('postgresql://root:root@localhost:5432/ny_taxi')
Solutions:
Port Change:
- Change the port from 5432 to another port (e.g., 5431).
- Example: Change
5432:5432to5431:5432.
User Change:
- Change
POSTGRES_USER=roottoPGUSER=postgres.
- Change
Docker Solution:
- Run
docker compose down. - Remove the folder containing the Postgres volume.
- Run
docker compose upagain.
- Run
Additional Resources:
For more details, refer to this Stack Overflow discussion.
# Postgres - OperationalError: (psycopg2.OperationalError) connection to server at "localhost" (::1), port 5432 failed: FATAL: database "ny_taxi" does not exist
OperationalError: (psycopg2.OperationalError) connection to server at "localhost" (::1), port 5432 failed: FATAL: database "ny_taxi" does not exist
Make sure PostgreSQL is running. You can check that by running:
docker ps
Solution:
- If you have PostgreSQL software installed on your computer previously, consider building your instance on a different port, such as 8080, instead of 5432.
# Postgres - ModuleNotFoundError: No module named 'psycopg2'
Issue:
ModuleNotFoundError: No module named 'psycopg2'
Solution:
Install psycopg2-binary:
pip install psycopg2-binaryIf psycopg2-binary is already installed, update it:
pip install psycopg2-binary --upgradeOther methods if the above fails:
If the error persists, update conda:
conda update -n base -c defaults condaAlternatively, update pip:
pip install --upgrade pipReinstall psycopg:
- Uninstall the psycopg package.
- Update conda or pip.
- Reinstall psycopg using pip.
If an error shows about
pg_confignot being found, install PostgreSQL:On Mac, use:
brew install postgresql
# Postgres: "Column does not exist" but it actually does (Pyscopg2 error in MacBook Pro M2)
In join queries, if you mention the column name directly or enclose it in single quotes, you'll encounter an error saying "column does not exist".
Solution: Enclose the column names in double quotes, and it will work correctly.
# pgAdmin: Create server dialog does not appear
pgAdmin has a new version. The create server dialog may not appear. Try using Register -> Server instead.
# pgAdmin - Blank/white screen after login (browser)
Using GitHub Codespaces in the browser resulted in a blank screen after logging into pgAdmin (running in a Docker container). The terminal of the pgAdmin container was showing the following error message:
CSRFError: 400 Bad Request: The referrer does not match the host.
Solution #1:
As recommended in the following issue: GitHub Issue #5432, setting the following environment variable solved it:
docker run --rm -it \
-e PGADMIN_DEFAULT_EMAIL="admin@admin.com" \
-e PGADMIN_DEFAULT_PASSWORD="root" \
-e PGADMIN_CONFIG_WTF_CSRF_ENABLED="False" \
-p "8080:80" \
--name pgadmin \
--network=pg-network \
dpage/pgadmin4:8.2
Solution #2:
Using the locally installed VSCode to display GitHub Codespaces. When using GitHub Codespaces in the locally installed VSCode (opening a Codespace or creating/starting one), this issue did not occur.
# pgAdmin - Can not access/open the PgAdmin address via browser
I am using a Mac Pro device and connect to the GCP Compute Engine via Remote SSH - VSCode. But when trying to run the PgAdmin container via docker run or docker compose, I couldn't access the PgAdmin address via my browser. After modifications, I was able to access it.
Solution #1:
Modify the docker run command:
docker run --rm -it \
-e PGADMIN_DEFAULT_EMAIL="admin@admin.com" \
-e PGADMIN_DEFAULT_PASSWORD="pgadmin" \
-e PGADMIN_CONFIG_WTF_CSRF_ENABLED="False" \
-e PGADMIN_LISTEN_ADDRESS=0.0.0.0 \
-e PGADMIN_LISTEN_PORT=5050 \
-p 5050:5050 \
--network=de-zoomcamp-network \
--name pgadmin-container \
--link postgres-container \
-t dpage/pgadmin4
Solution #2:
Modify the docker-compose.yaml configuration and use the docker compose up command:
pgadmin:
image: dpage/pgadmin4
container_name: pgadmin-container
environment:
- PGADMIN_DEFAULT_EMAIL=admin@admin.com
- PGADMIN_DEFAULT_PASSWORD=pgadmin
- PGADMIN_CONFIG_WTF_CSRF_ENABLED=False
- PGADMIN_LISTEN_ADDRESS=0.0.0.0
- PGADMIN_LISTEN_PORT=5050
volumes:
- "./pgadmin_data:/var/lib/pgadmin/data"
ports:
- "5050:5050"
networks:
- de-zoomcamp-network
depends_on:
- postgres-container
# pgAdmin: How to Persist pgAdmin Configurations
To keep pgAdmin settings after restarting the container, follow these steps:
Create the directory for pgAdmin data:
mkdir -p /path/to/pgadmin-dataAssign ownership to pgAdmin's user (ID 5050):
sudo chown -R 5050:5050 /path/to/pgadmin-dataSet the appropriate permissions:
sudo chmod -R 755 /path/to/pgadmin-data
# pgAdmin - Unable to connect to server: [Errno -3] Try again
This error occurs when connecting pgAdmin with Docker Postgres. In the tutorial, the pgAdmin server creation under Connection > Host name/address uses pg-database and results in the above-mentioned error when saved.
Solution 1:
Verify that both containers are connected to
pg-network:docker network inspect pg-networkIf the Docker Postgres container is not connected, connect it to
pg-network:docker network connect pg-network postgresContainer_nameRetry the connection. If the error persists, instead of using
pg-databaseunder Connection > Host name/address, try using the IP Address:- Use the IP address of the
postgresContainer_namecontainer (e.g.,172.19.0.3) in the pgAdmin configuration instead of the container name orpg-database.
- Use the IP address of the
# Python - ModuleNotFoundError: No module named 'pysqlite2'
ImportError: DLL load failed while importing _sqlite3: The specified module could not be found.
ModuleNotFoundError: No module named 'pysqlite2'
The issue may arise due to the absence of sqlite3.dll in the path ".\Anaconda\Dlls\".
To resolve the issue:
- Copy the
sqlite3.dllfile from\Anaconda3\Library\bin. - Paste the file into the
".\Anaconda\Dlls\"directory.
This solution applies if you are using Anaconda.
# Python: Ingestion with Jupyter notebook - missing 100000 records
If you follow the video 1.2.2 - Ingesting NY Taxi Data to Postgres and execute the same steps, you will ingest all the data (~1.3 million rows) into the table yellow_taxi_data. However, running the whole script in the Jupyter notebook for a second time from top to bottom will result in missing the first chunk of 100,000 records. This occurs because a call to the iterator appears before the while loop, leading to the second chunk being ingested first.
Solution:
- Remove the cell
df=next(df_iter)located higher up in the notebook than the while loop. - Ensure the first
w(df_iter)call is within the while loop.
📔 Note: The notebook is used to test the code and is not intended to be run top to bottom. The logic is organized in a later step when inserted into a .py file for the pipeline.
# IPython - Pandas parsing dates with "read_csv"
Pandas can interpret "string" column values as "datetime" directly when reading the CSV file using pd.read_csv with the parse_dates parameter. This can include a list of column names or column indices, eliminating the need for conversion afterward.
Reference: pandas.read_csv documentation
Example from Week 1:
import pandas as pd
df = pd.read_csv(
'yellow_tripdata_2021-01.csv',
nrows=100,
parse_dates=['tpep_pickup_datetime', 'tpep_dropoff_datetime']
)
df.info()
Output:
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 100 entries, 0 to 99
Data columns (total 18 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 VendorID 100 non-null int64
1 tpep_pickup_datetime 100 non-null datetime64[ns]
2 tpep_dropoff_datetime 100 non-null datetime64[ns]
3 passenger_count 100 non-null int64
4 trip_distance 100 non-null float64
5 RatecodeID 100 non-null int64
6 store_and_fwd_flag 100 non-null object
7 PULocationID 100 non-null int64
8 DOLocationID 100 non-null int64
9 payment_type 100 non-null int64
10 fare_amount 100 non-null float64
11 extra 100 non-null float64
12 mta_tax 100 non-null float64
13 tip_amount 100 non-null float64
14 tolls_amount 100 non-null float64
15 improvement_surcharge 100 non-null float64
16 total_amount 100 non-null float64
17 congestion_surcharge 100 non-null float64
dtypes: datetime64[ns](2), float64(9), int64(6), object(1)
memory usage: 14.2+ KB
# Python: Python can't ingest data from the GitHub link provided using curl
os.system(f"curl -LO {url} -o {csv_name}")
# Python: Pandas can read *.csv.gzip
When a CSV file is compressed using Gzip, it is saved with a ".csv.gz" file extension. This file type is also known as a Gzip compressed CSV file. To read a Gzip compressed CSV file using Pandas, you can use the read_csv() function.
Here is an example of how to read a Gzip compressed CSV file using Pandas:
import pandas as pd
df = pd.read_csv('file.csv.gz',
compression='gzip',
low_memory=False)
# Python: How to iterate through and ingest parquet file
Contrary to pandas’ read_csv method, there’s no simple way to iterate through and set chunksize for parquet files. We can use PyArrow (Apache Arrow Python bindings) to resolve that.
import pyarrow.parquet as pq
from sqlalchemy import create_engine
import time
output_name = "https://d37ci6vzurychx.cloudfront.net/trip-data/yellow_tripdata_2021-01.parquet"
parquet_file = pq.ParquetFile(output_name)
parquet_size = parquet_file.metadata.num_rows
engine = create_engine(f'postgresql://{user}:{password}@{host}:{port}/{db}')
table_name = "yellow_taxi_schema"
# Clear table if exists
pq.read_table(output_name).to_pandas().head(n=0).to_sql(name=table_name, con=engine, if_exists='replace')
# Default (and max) batch size
index = 65536
for i in parquet_file.iter_batches(use_threads=True):
t_start = time.time()
print(f'Ingesting {index} out of {parquet_size} rows ({index / parquet_size:.0%})')
i.to_pandas().to_sql(name=table_name, con=engine, if_exists='append')
index += 65536
t_end = time.time()
print(f'\t- it took %.1f seconds' % (t_end - t_start))
# Python - SQLAlchemy - ImportError: cannot import name 'TypeAliasType' from 'typing_extensions'.
The following error occurs during the execution of a Jupyter notebook cell:
from sqlalchemy import create_engine
Solution:
The issue can be resolved by ensuring the version of the Python module typing_extensions is 4.6.0 or later. You can update it using either Conda or pip:
Using Conda:
conda update typing_extensionsUsing pip:
pip install --upgrade typing_extensions
For more details, you can refer to the changelog for typing_extensions 4.6.0.
# Python - SQLALchemy - TypeError 'module' object is not callable
When using create_engine('postgresql://root:root@localhost:5432/ny_taxi'), you may encounter the error:
TypeError: 'module' object is not callable
Use the correct connection string syntax:
conn_string = "postgresql+psycopg://root:root@localhost:5432/ny_taxi"
engine = create_engine(conn_string)
# Python: SQLAlchemy - ModuleNotFoundError: No module named 'psycopg2'.
Error raised during the Jupyter Notebook cell execution:
engine = create_engine('postgresql://root:root@localhost:5432/ny_taxi')
Solution:
Install the Python module psycopg2. It can be installed using Conda or pip:
- Using Conda:
conda install psycopg2 - Using pip:
pip install psycopg2
# Python - SQLAlchemy: NoSuchModuleError: Can't load plugin: sqlalchemy.dialects:postgresql.psycopg
Error raised during the Jupyter notebook’s cell execution:
conn_string = "postgresql+psycopg://root:root@localhost:5432/ny_taxi"
engine = create_engine(conn_string)
Solution:
We had a scenario of a virtual environment (created by PyCharm) being run on top of another virtual environment (on conda). The solution was:
Remove the
.venv.Create a new virtual environment with conda:
conda create -n pyingest python=3.12Install the required dependencies:
pip install pandas sqlalchemy psycopg2-binary jupyterlabRe-execute the code.
For psycopg2, the connection string should be:
postgresql+psycopg2://{db_user}:{db_password}@{db_host}:{db_port}/{db_name}
Reference - Kayla Tinker 1/14/25
# Python - SQLAlchemy - read_sql_query() throws "'OptionEngine' object has no attribute 'execute'"
First, check the versions of SQLAlchemy and Pandas to ensure they are both up-to-date. You can upgrade them using pip or conda if needed.
Then, try to wrap the query using text:
from sqlalchemy import text
query = text("SELECT * FROM tbl")
df = pd.read_sql_query(query, conn)
# GCP: Static vs Ephemeral IP / Setting up static IP for VM
When you set up a VM in Google Cloud Platform (GCP), it initially uses an ephemeral IP address, which changes each time you start or stop the VM. If you need a consistent IP for your configuration file, you should set up a static IP address.
Steps to Set Up a Static IP Address
- Navigate to VPC Network > IP addresses in the GCP console.
- Allocate a new static IP address.
- Attach the static IP to your VM instance.
Note: You are charged for a static IP if it is not allocated to a specific VM, so make sure it is attached to avoid extra fees.
For detailed instructions, consult the GCP documentation.
# GCP: Unable to add Google Cloud SDK PATH to Windows
Issue
Windows error:
The installer is unable to automatically update your system PATH. Please add C:\tools\google-cloud-sdk\bin
Solution
If you encounter this error frequently, consider the following steps:
Add Gitbash to Windows Path:
- Using Conda:
- Download the Anaconda Navigator.
- During installation, check the box to add Conda to the path (even though it's not recommended).
- Using Conda:
Install Git Bash:
- If not installed, install Git Bash.
- If installed, consider reinstalling it.
- During installation, ensure you check:
- Add GitBash to Windows Terminal
- Use Git and optional Unix tools from the command prompt
Setup Git Bash:
Open Git Bash and type the following command:
conda init bashThis will modify your bash profile.
Set Gitbash as Default Terminal:
- Open the Windows Terminal.
- Go to settings.
- Change the default profile from Windows PowerShell to Git Bash.
By following these steps, you should be able to add the Google Cloud SDK path to your system on Windows without issues.
# GCP: Project creation failed: HttpError accessing … Requested entity already exists
When creating a project in GCP, you may encounter the following error:
WARNING: Project creation failed: HttpError accessing cloudresourcemanager.googleapis.com: response: {
'content-type': 'application/json; charset=UTF-8',
'status': 409
}, content {
"error": {
"code": 409,
"message": "Requested entity already exists",
"status": "ALREADY_EXISTS"
}
}
Explanation
This error occurs when the project ID you are trying to use is already taken. Project IDs are unique across all GCP projects. If any user ever had a project with that ID, you cannot use it.
Solution
- Choose a different, more unique project ID. Avoid common names like
testprojectas they are likely to be already in use.
For more details, refer to the discussion: Stack Overflow
# GCP: The project to be billed is associated with an absent billing account
If you receive the error:
Error 403: The project to be billed is associated with an absent billing account., accountDisabled
It is most likely because you did not enter your project ID correctly. The value you enter should be unique to your project. You can find this value on your GCP Dashboard when you log in.
Another possibility is that you have not linked your billing account to your current project.
# GCP: OR-CBAT-15 ERROR Google cloud free trial account
If Google refuses your credit/debit card, try using a different one. For instance, a card from Kaspi (Kazakhstan) might not work, but a card from TBC (Georgia) does.
Unfortunately, support assistance might not be highly effective in resolving this issue.
Additionally, a Pyypl web-card can be a viable alternative.
ny-rides.json
# GCP: Where can I find the “ny-rides.json” file?
The ny-rides.json is your private file in Google Cloud Platform (GCP). Here’s how to find it:
- Navigate to GCP and select the project with your instance.
- Go to IAM & Admin.
- Select the Service Accounts tab.
- Click the Keys tab.
- Add a key, choosing JSON as the key type, then click Create.
Note: Once in the Service Accounts, click the email associated to access the KEYS tab where you can add a key as a JSON key type.
# GCP: "Failed to load" when accessing Compute Engine’s metadata section (e.g., to add a SSH key)
You likely didn’t enable the Compute Engine API.
# GCP: Do I need to delete my instance in Google Cloud?
In this lecture, Alexey deleted his instance in Google Cloud. Do I have to do it?
No, do not delete your instance in Google Cloud Platform. Otherwise, you will have to set it up again for the week 1 readings.
# GCP: SSH public key error - multiple users / usernames
Initially, I could not SSH into my VM from my Windows laptop. I thought it was because I did not follow the tutorial exactly. Instead of generating the SSH key using MINGW/git bash with the Linux-style command, I did it in Command Prompt using the Windows-style command. I kept getting a public key error.
Permanent Solution:
It turns out it wasn’t an issue with the key generation at all! The problem was with the username. I had given my SSH key a different username than what appeared in my VM (my Google account username). So, I had been trying to log in with googleacctuser@[ipaddr] instead of mySSHuser@[ipaddr]. Here's how I resolved it:
- Retraced my steps to check the SSH key setup in the GCP console, where it showed the user and SSH key.
- Changed the username to the correct one (googleacctuser) in my config file.
- Updated the config file and used
mySSHuserto log in.
Now, the issue was that I had created two users. I made all the installations and permissions on googleacctuser, not accessible from mySSHuser. Since I didn't need mySSHuser, I edited the SSH key to change the username at the end and updated the GCP console and config file accordingly.
Then, I planned to delete the mySSHuser account in the VM terminal to keep things clean (though I got a bit attached, so I skipped this).
Temporary Solution:
Before figuring out my issue, I used a shortcut by SSH'ing into the VM in the browser, which worked nicely for a while. But eventually, I needed to use VSCode.
# GCP: Virtual Machine (VM) Size, Slow, Clean Up
If you are progressing through the course and find that your VM is starting to become slow, you can run the following commands to inspect and detect areas where you can improve:
Recommended VM Size
- Start with a 60GB machine. A 30GB machine may not be sufficient, as you might need to restart the project with a larger size.
Commands to Inspect the Health of Your VM
System Resource Usage
top htopShows real-time information about system resource usage, including CPU, memory, and processes.
free -hDisplays information about system memory usage and availability.
df -hShows disk space usage of file systems.
du -h <directory>Displays disk usage of a specific directory.
Running Processes
ps auxLists all running processes along with detailed information.
Network
ifconfig ip addr showShows network interface configuration.
netstat -tulnDisplays active network connections and listening ports.
Hardware Information
lscpuDisplays CPU information.
lsblkLists block devices (disks and partitions).
lshwLists hardware configuration.
User and Permissions
whoShows who is logged on and their activities.
wDisplays information about currently logged-in users and their processes.
Package Management
apt list --installedLists installed packages (for Ubuntu and Debian-based systems).
# Billing: Billing account has not been enabled for this project. But you’ve done it indeed!
If you’ve got the error:
Error: Error updating Dataset "projects/<your-project-id>/datasets/demo_dataset": googleapi: Error 403: Billing has not been enabled for this project. Enable billing at console.cloud.google.com. The default table expiration time must be less than 60 days, billingNotEnabled
but you’ve set your billing account, try disabling billing for the project and enabling it again. This method has been successful for others.
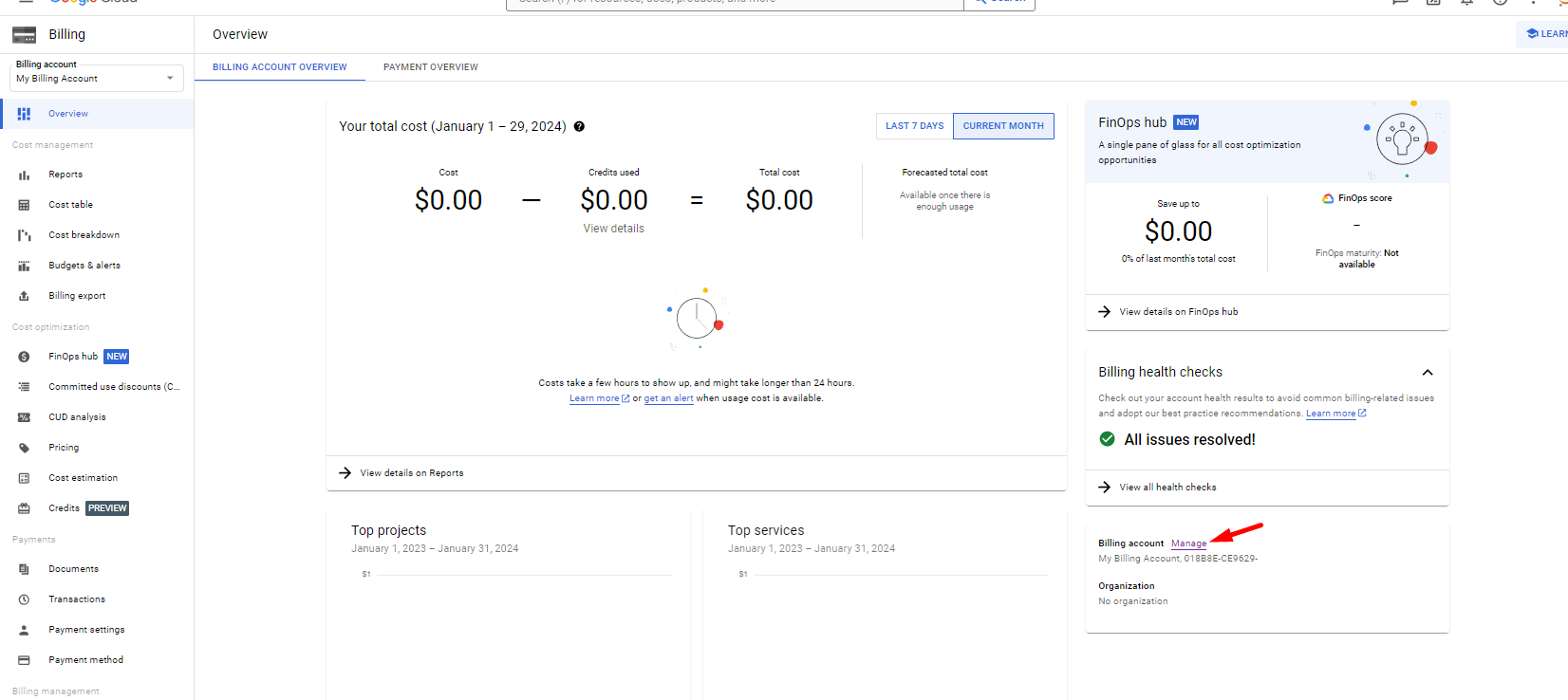
# GCP - Windows Google Cloud SDK install issue:
If you are encountering installation trouble with the Google Cloud SDK on Windows and receiving the following error:
These credentials will be used by any library that requests Application Default Credentials (ADC).
WARNING:
Cannot find a quota project to add to ADC. You might receive a "quota exceeded" or "API not enabled" error. Run $ gcloud auth application-default set-quota-project to add a quota project.
Try these steps:
- Reinstall the SDK using the unzip file "install.bat".
- Check the installation by running
gcloud version. - Run
gcloud initto set up your project. - Execute
gcloud auth application-default login.
For detailed instructions, refer to the following guide: Windows SDK Installation Guide
# GCP: I cannot get my Virtual Machine to start because GCP has no resources.
- Click on your VM.
- Create an image of your VM.
- On the page of the image, tell GCP to create a new VM instance via the image.
- On the settings page, change the location.
# GCP VM: Is it necessary to use a GCP VM? When is it useful?
The reason this video about the GCP VM exists is that many students had problems configuring their environment. You can use your own environment if it works for you.
Advantages of using your own environment include:
- Commit Changes: If you are working in a GitHub repository, you will be able to commit changes directly. In the VM, the repo is cloned via HTTPS, so it is not possible to commit directly, even if you are the owner of the repo.
# GCP VM - mkdir: cannot create directory ‘.ssh’: Permission denied
If you encounter an error while trying to create a directory:
User1@DESKTOP-PD6UM8A MINGW64 /
$ mkdir .ssh
mkdir: cannot create directory ‘.ssh’: Permission denied
This error occurs because you are attempting to create the directory in the root folder (/).
To resolve this, create the directory in your home directory instead. Use the following steps:
Navigate to your home directory using:
cd ~Create the
.sshdirectory:mkdir .ssh
For further guidance, watch this video.
# GCP VM: Error while saving the file in VM via VS Code
Failed to save '<file>': Unable to write file 'vscode-remote://ssh-remote+de-zoomcamp/home/<user>/data_engineering_course/week_2/airflow/dags/<file>' (NoPermissions (FileSystemError): Error: EACCES: permission denied, open '/home/<user>/data_engineering_course/week_2/airflow/dags/<file>')
To resolve this issue, you need to change the owner of the files you are trying to edit via VS Code. Follow these steps:
Connect to your VM using SSH.
Run the following command to change the ownership:
sudo chown -R <user> <path to your directory>
# GCP VM: VM connection request timeout
Question: I connected to my VM perfectly fine last week (SSH) but when I tried again this week, the connection request keeps timing out.
Answer:
Start Your VM: Make sure the VM is running in your GCP console.
Update External IP:
- Copy its External IP once the VM is running.
- Update your SSH configuration file with this IP.
Edit SSH Config:
cd ~/.ssh code configThis command opens the config file in VSCode for editing.
# GCP VM: connect to host port 22 no route to host
Go to edit your VM.
Navigate to the Automation section.
Add the following Startup script:
#!/bin/bash sudo ufw allow sshStop and Start the VM.
# GCP VM: Port forwarding from GCP without using VS Code
You can easily forward the ports of pgAdmin, PostgreSQL, and Jupyter Notebook using the built-in tools in Ubuntu without any additional client:
On the VM machine:
- Launch Docker and Jupyter Notebook in the correct folder using:
docker-compose up -d jupyter notebook
- Launch Docker and Jupyter Notebook in the correct folder using:
From the local machine:
- Execute:
ssh -i ~/.ssh/gcp -L 5432:localhost:5432 username@external_ip_of_vm - Execute the same command for ports 8080 and 8888.
- Execute:
Accessing Applications Locally:
- For pgAdmin, open a browser and go to
localhost:8080. - For Jupyter Notebook, open a browser and go to
localhost:8888.- If you encounter issues with credentials, you may need to copy the link with the access token from the terminal logs on the VM when you launched the Jupyter Notebook.
- For pgAdmin, open a browser and go to
Forwarding Both pgAdmin and PostgreSQL:
- Use:
ssh -i ~/.ssh/gcp -L 5432:localhost:5432 -L 8080:localhost:8080 modito@35.197.218.128
- Use:
# GCP gcloud + MS VS Code - gcloud auth hangs
If you are using MS VS Code and running gcloud in WSL2, when you first try to login to GCP via the gcloud CLI with gcloud auth application-default login, you may encounter an issue where a message appears and nothing happens:
There might be a prompt asking if you want to open it via a browser. If you click on it, it will open a page with an error message:
Solution:
- Hover over the long link.
Ctrl + Clickthe long link.- Click "Configure Trusted Domains here."
- A popup will appear; pick the first or second entry.

Next time you run gcloud auth, the login page should pop up via the default browser without issues.
# Terraform - Error: Failed to query available provider packages │ Could not retrieve the list of available versions for provider hashicorp/google: could not query │ provider registry for registry.terrafogorm.io/hashicorp/google: the request failed after 2 attempts, │ please try again later
This error typically occurs due to internet connectivity issues. Terraform is unable to access the online registry.
Solution:
- Check your VPN/Firewall settings.
- Clear cookies or restart your network.
- Run
terraform initagain after addressing the connection issues.
# Terraform: Error: Post "[storage.googleapis.com](https://storage.googleapis.com/storage/v1/b?alt=json&prettyPrint=false&project=coherent-ascent-379901): oauth2: cannot fetch token: Post "[oauth2.googleapis.com](https://oauth2.googleapis.com/token): dial tcp 172.217.163.42:443: i/o timeout
The issue was related to network restrictions, as Google is not accessible in my country. I used a VPN and discovered that the terminal program does not automatically follow the system proxy, requiring separate proxy configuration settings.
Solution:
- Open an Enhanced Mode in your VPN application, such as Clash.
- Run
terraform applyagain.
If you encounter this issue, consult your VPN provider for assistance with configuration.
# Terraform: Install for WSL
You can configure Terraform on Windows 10 using the Linux Subsystem (WSL) by following this guide: Configuring Terraform on Windows 10 Linux Subsystem.
# Terraform: Error acquiring the state lock
For more information, you can refer to the following issue on GitHub:
# Terraform: Error 400 Bad Request. Invalid JWT Token on WSL.
When running:
terraform apply
on WSL2, you might encounter the following error:
Error: Post "https://storage.googleapis.com/storage/v1/b?alt=json&prettyPrint=false&project=<your-project-id>": oauth2: cannot fetch token: 400 Bad Request
Response: {"error":"invalid_grant","error_description":"Invalid JWT: Token must be a short-lived token (60 minutes) and in a reasonable timeframe. Check your iat and exp values in the JWT claim."}
This issue occurs due to potential time desynchronization on your machine, affecting JWT computation.
To fix this, run the following command to synchronize your system time:
sudo hwclock -s
# Terraform - Error 403 : Access denied
│ Error: googleapi: Error 403: Access denied., forbidden
Your $GOOGLE_APPLICATION_CREDENTIALS might not be pointing to the correct file. Try the following steps:
Set the correct path for your credentials:
export GOOGLE_APPLICATION_CREDENTIALS=~/.gc/YOUR_JSON.jsonActivate the service account:
gcloud auth activate-service-account --key-file $GOOGLE_APPLICATION_CREDENTIALS
# Terraform: Do I need to make another service account for Terraform before I get the keys (.json file)?
One service account is enough for all the services/resources you'll use in this course. After you get the file with your credentials and set your environment variable, you should be good to go.
# Terraform: Where can I find the Terraform 1.1.3 Linux (AMD 64)?
# Terraform: Terraform initialized in an empty directory! The directory has no Terraform configuration files. You may begin working with Terraform immediately by creating Terraform configuration files.
This error occurs when terraform init is run outside the working directory.
To resolve this issue:
- Navigate to the working directory that contains your Terraform configuration files.
- Run the
terraform initcommand inside the correct directory.
Make sure your configuration files (e.g., .tf files) are present in the directory before running the command.
# Terraform - Error creating Dataset: googleapi: Error 403: Request had insufficient authentication scopes
The error:
Error: googleapi: Error 403: Access denied., forbidden
Error: Error creating Dataset: googleapi: Error 403: Request had insufficient authentication scopes.
Solution:
Verify your credentials by running:
echo $GOOGLE_APPLICATION_CREDENTIALS echo $?Ensure you have set the
GOOGLE_APPLICATION_CREDENTIALSenvironment variable correctly, as demonstrated in the environment setup video in week 1:export GOOGLE_APPLICATION_CREDENTIALS="<path/to/your/service-account-authkeys>.json"
# stoTerraform - Error creating Bucket: googleapi: Error 403: Permission denied to access ‘storage.buckets.create’
The error:
Error: googleapi: Error 403: terraform-trans-campus@trans-campus-410115.iam.gserviceaccount.com does not have storage.buckets.create access to the Google Cloud project. Permission 'storage.buckets.create' denied on resource (or it may not exist)., forbidden
The solution:
You have to declare the project name as your Project ID, not your Project name, available on the GCP console Dashboard.
# Terraform: google provider requires credentials.
To ensure the sensitivity of the credentials file, use the following configuration:
provider "google" {
project = var.projectId
credentials = file("${var.gcpkey}")
#region = var.region
zone = var.zone
}
# Terraform: Teardown of BigQuery Dataset
When running terraform destroy, the following error can occur:
Do you really want to destroy all resources?
Terraform will destroy all your managed infrastructure, as shown above.
There is no undo. Only 'yes' will be accepted to confirm.
Enter a value: yes
google_bigquery_dataset.homework_dataset: Destroying... [id=projects/terraform-demo-449214/datasets/homework_dataset]
╷
│ Error: Error when reading or editing Dataset: googleapi: Error 400: Dataset terraform-demo-449214:homework_dataset is still in use, resourceInUse
This is because the dataset is still in use by a table. To delete the dataset, set the delete_contents_on_destroy property to true in the main.tf file.
# SQL: SELECT * FROM zones_taxi WHERE Zone='Astoria Zone'; Error Column Zone doesn't exist
For this issue, you can use the following solution:
SELECT * FROM zones AS z WHERE z."Zone" = 'Astoria Zone';
Columns that start with uppercase sometimes need to be enclosed in double quotes.
Additionally, check your dataset for the existence of 'Astoria Zone'. You might find only 'Astoria':
SELECT * FROM zones AS z WHERE z."Zone" = 'Astoria';
# SQL: SELECT Zone FROM taxi_zones Error Column Zone doesn't exist
It is inconvenient to use quotation marks all the time, so it is better to put the data in the database all in lowercase. In Pandas, after:
import pandas as pd
df = pd.read_csv('taxi+_zone_lookup.csv')
Add the row:
df.columns = df.columns.str.lower()
# CURL: curl: (6) Could not resolve host: output.csv
os.system(f"curl {url} --output {csv_name}")
# SSH Error: ssh: Could not resolve hostname linux: Name or service not known
To resolve this, ensure that your config file is in C/User/Username/.ssh/config.
# 'pip' is not recognized as an internal or external command, operable program or batch file.
If you use Anaconda (recommended for the course), it comes with pip, so the issue is probably that Anaconda’s Python is not on the PATH.
For Linux and MacOS:
Open a terminal.
Find the path to your Anaconda installation. This is typically
~/anaconda3or~/opt/anaconda3.Add Anaconda to your PATH with the command:
export PATH="/path/to/anaconda3/bin:$PATH"To make this change permanent, add the command to your
.bashrc(Linux) or.bash_profile(MacOS) file.
On Windows, using Git Bash:
Locate your Anaconda installation. The default path is usually
C:\Users\[YourUsername]\Anaconda3.Convert the Windows path to a Unix-style path for Git Bash, e.g.,
C:\Users\[YourUsername]\Anaconda3becomes/c/Users/[YourUsername]/Anaconda3.Add Anaconda to your PATH with the command:
export PATH="/c/Users/[YourUsername]/Anaconda3/:/c/Users/[YourUsername]/Anaconda3/Scripts/$PATH"To make this change permanent, add the command to your
.bashrcfile in your home directory.Refresh your environment with the command:
source ~/.bashrc
For Windows (without Git Bash):
- Right-click on 'This PC' or 'My Computer' and select 'Properties'.
- Click on 'Advanced system settings'.
- In the System Properties window, click on 'Environment Variables'.
- In the Environment Variables window, select the 'Path' variable in the 'System variables' section and click 'Edit'.
- In the Edit Environment Variable window, click 'New' and add the path to your Anaconda installation (typically
C:\Users\[YourUsername]\Anaconda3andC:\Users\[YourUsername]\Anaconda3\Scripts). - Click 'OK' in all windows to apply the changes.
After adding Anaconda to the PATH, you should be able to use pip from the command line. Remember to restart your terminal (or command prompt in Windows) to apply these changes.
# Error: error starting userland proxy: listen tcp4 0.0.0.0:8080: bind: address already in use
Resolution: You need to stop the service using the port.
Run the following:
sudo kill -9 `sudo lsof -t -i:<port>`
Replace <port> with 8080 in this case. This will free up the port for use.
# Anaconda to PIP
To get a pip-friendly requirements.txt file from Anaconda, use the following steps:
- Install pip in your Anaconda environment:
conda install pip - Generate the
requirements.txtfile:pip list --format=freeze > requirements.txt
Note:
conda list -d > requirements.txtwill not work.pip freeze > requirements.txtmay give odd pathing.
# Jupyter: Install, open Jupyter and convert Jupyter notebook to Python script
Install and Open Jupyter Notebook
To install Jupyter Notebook, run:
pip install jupyter
To open Jupyter Notebook, use:
python3 -m notebook
Convert Jupyter Notebook to Python Script
First, ensure nbconvert is installed and upgraded:
pip install nbconvert --upgrade
Then, convert a Jupyter Notebook to a Python script with the following command:
python3 -m jupyter nbconvert --to=script upload-data.ipynb
# Alternative way to convert Jupyter notebook to Python script (via jupytext)
If you keep getting errors with nbconvert after executing:
jupyter nbconvert --to script <your_notebook.ipynb>
You could try converting your Jupyter notebook using another tool called Jupytext. Jupytext is an excellent tool for converting Jupyter Notebooks to Python scripts, similar to nbconvert.
Install Jupytext
pip install jupytextConvert your Notebook to a Python script
jupytext --to py <your_notebook.ipynb>
Module 2: Workflow Orchestration
# SSH error in VS Code - “Could not establish connection to "de-zoomcamp": Permission denied (publickey).”
If you are using Windows, try the following steps to resolve the error:
Copy the
.sshfolder from the Linux file path to Windows.In the
configfile, use:IdentityFile C:\Users\<username>\.ssh\gcpInstead of:
IdentityFile ~/.ssh/gcpEnsure the private key file located at
C:\Users\<username>\.ssh\gcphas an extra line at the end:
# Where are the FAQ questions from the previous cohorts for the orchestration module?
# How do I launch Kestra?
To launch Kestra, follow these instructions:
For Linux
Start Docker with the following command:
docker run \
--pull=always \
--rm \
-it \
-p 8080:8080 \
--user=root \
-v /var/run/docker.sock:/var/run/docker.sock \
-v /tmp:/tmp \
kestra/kestra:latest server local
Once it is running, you can log in to the dashboard at localhost:8080.
For Windows
Refer to the Kestra GitHub repository for detailed instructions: https://github.com/kestra-io/kestra
Sample docker-compose for Kestra:
kestra:
build: .
image: kestra/kestra:latest
container_name: kestra
user: "0:0"
environment:
DOCKER_HOST: tcp://host.docker.internal:2375 # for Windows
KESTRA_CONFIGURATION: |
kestra:
repository:
type: h2
queue:
type: memory
storage:
type: local
local:
basePath: /app/storage
tasks:
tmp-dir:
path: /app/tmp
plugins:
repositories:
- id: central
type: maven
url: [repo.maven.apache.org](https://repo.maven.apache.org/maven2)
definitions:
- io.kestra.plugin.core:core:latest
- io.kestra.plugin.scripts:python:1.3.4
- io.kestra.plugin.http:http:latest
KESTRA_TASKS_TMP_DIR_PATH: /app/tmp
ports:
- "8080:8080"
volumes:
- //var/run/docker.sock:/var/run/docker.sock # Windows path
- /yourpath/.dbt:/app/.dbt
- /yourpath/kestra/plugins:/app/plugins
- /yourpath/kestra/workflows:/app/workflows
- /yourpath/kestra/storage:/app/storage
- /yourpath//kestra/tmp:/app/tmp
- /yourpath//dbt_prj:/app/workflows/dbt_project
- /yourpath//my-creds.json:/app/.dbt/my-creds.json
command: server standalone
# docker: Error response from daemon: mkdir C:\Program Files\Git\var: Access is denied.
Description:
When running the following Docker command in Bash with Docker and WSL2 installed, you may encounter an error. Running Bash as admin will not resolve the issue:
docker run \
--pull=always \
--rm \
-it \
-p 8080:8080 \
--user=root \
-v /var/run/docker.sock:/var/run/docker.sock \
-v /tmp:/tmp \
kestra/kestra:latest server local
latest: Pulling from kestra/kestra
Digest: sha256:af02a309ccbb52c23ad1f1551a1a6db8cf0523cf7aac7c7eb878d7925bc85a62
Status: Image is up to date for kestra/kestra:latest
docker: Error response from daemon: mkdir C:\\Program Files\\Git\\var: Access is denied.
See 'docker run --help'.
To resolve this issue, run Command Prompt as an administrator and use the following command:
docker run \
--pull=always \
--rm \
-it \
-p 8080:8080 \
--user=root \
-v "/var/run/docker.sock:/var/run/docker.sock" \
-v "C:/Temp:/tmp" \
kestra/kestra:latest server local
After executing the command as described, the localhost should display the Kestra UI as expected.
# Error when running Kestra flow connecting to postgres
Error Message
org.postgresql.util.psqlexception the connection attempt failed due to this config on kestra flow -> jdbc:postgresql://host.docker.internal:5432/postgres-zoomcamp
Solution
- Replace
host.docker.internalwith the name of the service for Postgres in your Docker Compose file.
Additional Error Message
org.postgresql.util.PSQLException: The connection attempt failed. 2025-01-29 22:52:22.281 green_create_table The connection attempt failed. host.docker.internal
Analysis and Solution
If using Linux, the PostgreSQL database URL differs from the tutorial. Instead of
host.docker.internal, Linux users should use the service or container name for Postgres. For example, use:jdbc:postgresql://postgres:5432/kestraDouble-check the database name in your Docker Compose file. It might be different from the tutorial; for example,
kestrainstead ofpostgres-zoomcamp.
Reminder
- Ensure that the PostgreSQL database name in the Docker Compose matches what you configure in your flow.
# Adding a pgadmin service with volume mounting to the docker-compose:
I encountered an error where the localhost URL for pgAdmin would just hang (I chose localhost:8080 for my pgAdmin, and made kestra localhost:8090, personal preference).
The associated issue involved permissions. The resolution was to change the ownership of my local directory to the user "5050," which is pgAdmin. Unlike Postgres, pgAdmin requires explicit permission. Apparently, the Postgres user inside the Docker container creates the Postgres volume/dir, so it has permissions already.
This is a useful resource:
Stack Overflow: Permission denied /var/lib/pgadmin/sessions in Docker
# Running out of storage when using kestra with postgres on GCP VM
Running out of storage while trying to backfill. I realized my GCP VM only has 30GB of storage and I was using it up quickly. Here are a couple of suggestions for managing storage:
Clean up your GCP VM drive: Use the command below to identify what is taking up the most space:
sudo du -sh *(~1GB) For me, the Anaconda installer was consuming a lot of space. If you no longer need it, you can delete it:
rm -rf <anacondainstaller_fpath>(~3GB) Anaconda itself takes up a lot of space. You can’t delete it entirely if you need Python, but you can clean it up significantly:
conda clean --all -y
Clean up your Kestra files: Use a purge flow. You can find a generic example here:
https://kestra.io/docs/administrator-guide/purge
I wanted to perform the cleanup immediately, rather than waiting until the end of the month, so I adjusted the
endDateto"{{ now() }}"and removed the trigger block. You can also choose whether to remove FAILED state executions.Clean up your PostgreSQL database: You can manually delete tables in pgAdmin, or set up a workflow in Kestra for it. I found it easy to do manually.
# How can Kestra access service account credential?
Do not directly add the content of service account credential JSON in Kestra script, especially if you are pushing to GitHub. Follow the instruction to add the service account as a secret Configure Google Service Account.
When you need to use it in Kestra, you can pull it through {{ secret('GCP_SERVICE_ACCOUNT') }} in the pluginDefaults.
# Storage: Bucket Permission Denied Error when running the gcp_setup flow
When following the YouTube lesson and then running the gcp_setup flow, you might encounter a permission denied error.
To resolve this:
- Verify if the bucket already exists using the GCP console.
- If it exists, choose a different name for the bucket.
Note: The GCP bucket name must be unique globally across all buckets, as the bucket will be accessible by URL.
# Invalid dataset ID Error when running the gcp_setup flow
When following the YouTube lesson and then running the gcp_setup flow, the error occurs during the create_bq_dataset task.
The error is less clear, but it stems from using a dash in the dataset name. To resolve this, change the dataset name to something like "de_zoomcamp" to avoid using a dash. This should resolve the error.
# How do I properly authenticate a Google Cloud Service Account in Kestra?
Several authentication methods are available; here are some of the most straightforward approaches:
Method 1:
Update your docker-compose.yml file as needed.
Method 2:
Store the Service Account as a Secret
Run this command, specifying the correct path to yourservice-account.jsonfile and.env_encoded:# Example command: Adjust according to your environment base64 /path/to/service-account.json > .env_encodedModify
docker-compose.ymlto Include the Encoded Secrets
Insert the relevant configuration within yourdocker-compose.yml.Configure Kestra Plugin Defaults
This ensures all GCP tasks use the secret automatically.Verify it’s Working in a Testing GCP Workflow
Additional FAQs:
Question: How do I update the Service Account key?
Answer: Generate a new key, re-run the Base64 command, and restart Kestra.
Question: Why use secrets instead of embedding the JSON key in the task?
Answer: Secrets prevent credential exposure and make workflows easier to manage.
Question: Can I apply this method to other GCP tasks?
Answer: Yes, all GCP plugins will automatically inherit the secret.
# Should I include my .env_encoded file in my .gitignore?
Yes, you should definitely include the .env_encoded file in your .gitignore file. Here's why:
Security: The
.env_encodedfile contains sensitive information, namely the base64 encoded version of your GCP Service Account key. Even though it's encoded, it's not secure to share this in a public repository as anyone can decode it back to the original JSON.Best Practices: It's common practice to avoid committing environment files or any files containing secrets to version control systems like Git. This prevents accidental exposure of sensitive data.
How to do it:
Add this line to your
.gitignore:.env_encoded
More on Security
Base64 encoding is easily reversible. Base64 is an encoding scheme, not an encryption method. It's designed to encode binary data into ASCII characters that can be safely transmitted over systems that are designed to deal with text. Here's why it's not secure for protecting sensitive information:
Reversibility: Base64 encoding simply translates binary data into a text string using a specific set of 64 characters. Decoding it back to the original data is straightforward and doesn't require any secret key or password.
Public Availability of Tools: Numerous online tools, software libraries, and command-line utilities exist that can decode base64 with just a few clicks or commands.
No Security: Since base64 encoding does not change or hide the actual content of the data, anyone with access to the encoded string can decode it back to the original data.
# Getting SIGILL in JRE when running latest kestra image on Mac M4 MacOS 15.2/3
SIGILL in Java Runtime Environment on MacOS M4
Add the following environment variable to your Kestra container: -e JAVA_OPTS="-XX:UseSVE=0":
docker run --rm -it \
--pull=always \
-p 8080:8080 \
--user=root \
-e JAVA_OPTS="-XX:UseSVE=0" \
-v /var/run/docker.sock:/var/run/docker.sock \
-v /tmp:/tmp \
kestra/kestra:latest server local
The same in a Docker Compose file:
services:
kestra:
image: kestra/kestra:latest
environment:
JAVA_OPTS: "-XX:UseSVE=0"
# taskid: yellow_create_table The connection attempt failed. Host.docker.internal
If you're using Linux, you might encounter "Connection Refused" errors when connecting to the Postgres DB from within Kestra. This is because host.docker.internal works differently on Linux.
To address this issue:
- Use the modified Docker Compose file mentioned in the "02-workflow-orchestration" README troubleshooting tips.
- Run both Kestra and its dedicated Postgres DB, along with the Postgres DB for exercises, all together using Docker Compose.
- Access the Postgres DB within Kestra by using the container name
postgres_zoomcampinstead ofhost.docker.internalinpluginDefaults.
Make sure to modify the pluginDefaults in the following files:
2_postgres_taxi_scheduled.yaml02_postgres_taxi.yaml
# Fix: Add extra_hosts for host.docker.internal on Linux
This update corrects the Docker Compose configuration to resolve the error when using the alias host.docker.internal on Linux systems. Since this alias does not resolve natively on Linux, the following entry was added to the affected container:
yaml
kestra:
image: kestra/kestra:latest
pull_policy: always
user: "root"
command: server standalone
volumes:
# Add volume configurations here
environment:
# Add environment variables here
ports:
# Add ports here
depends_on:
# Add dependencies here
extra_hosts:
- "host.docker.internal:host-gateway"
With this change, containers that need to access host services via host.docker.internal will be able to do so correctly. For inter-container communication within the same network, it is recommended to use the service name directly.
# Fix: Add extra_hosts for taskRunner in the dbt-build
To resolve the issue with host.docker.internal not being recognized on Linux, add the extraHosts configuration to the taskRunner in the dbt-build task:
taskRunner:
type: io.kestra.plugin.scripts.runner.docker.Docker
extraHosts:
- "host.docker.internal:host-gateway"
# Kestra: Don’t forget to set GCP_CREDS variable
If you plan on using Kestra with Google Cloud Platform, make sure you set up the GCP_CREDS that will be used in flows with "gcp" in their name.
To set it:
- Go to Namespaces and select "zoomcamp" if you are using the examples from the lessons.
- In the KV Store tab, create a new key as
GCP_CREDS. - Set the type to JSON and paste the content of the
.jsonfile with credentials for the service account created.
Module 3: Data Warehousing
# Kestra: Backfill showing getting executed but not getting results or showing up in executions
It seems to be a bug. The current fix is to remove the timezone from triggers in the script. More on this bug is here.
# Docker: Docker-compose takes infinitely long to install zip unzip packages for linux, which are required to unpack datasets.
To resolve the issue, you can try the following solutions:
Add the
-Yflag toapt-getto automatically agree to install additional packages.sudo apt-get install -y zip unzipUse the Python
ZipFilepackage, which is included in all modern Python distributions. This can bypass the need to installzipandunzippackages.from zipfile import ZipFile with ZipFile('file.zip', 'r') as zip_ref: zip_ref.extractall('destination_folder')
# GCS Bucket - error when writing data from web to GCS:
Make sure to use Nullable data types, such as Int64 when applicable.
# GCS Bucket - te table: Error while reading data, error message: Parquet column 'XYZ' has type INT which does not match the target cpp_type DOUBLE. File: gs://path/to/some/blob.parquet
Ultimately, when trying to ingest data into a BigQuery table, all files within a given directory must have the same schema.
When dealing with datasets, such as the FHV Datasets from 2019, you may encounter schema inconsistencies. For example, the files for '2019-05' and '2019-06' have the columns "PUlocationID" and "DOlocationID" as integers, while for the period of '2019-01' through '2019-04', the same columns are defined as floats.
When importing these files as Parquet to BigQuery, the first file will define the table schema. All subsequent files must have the same schema to append data correctly.
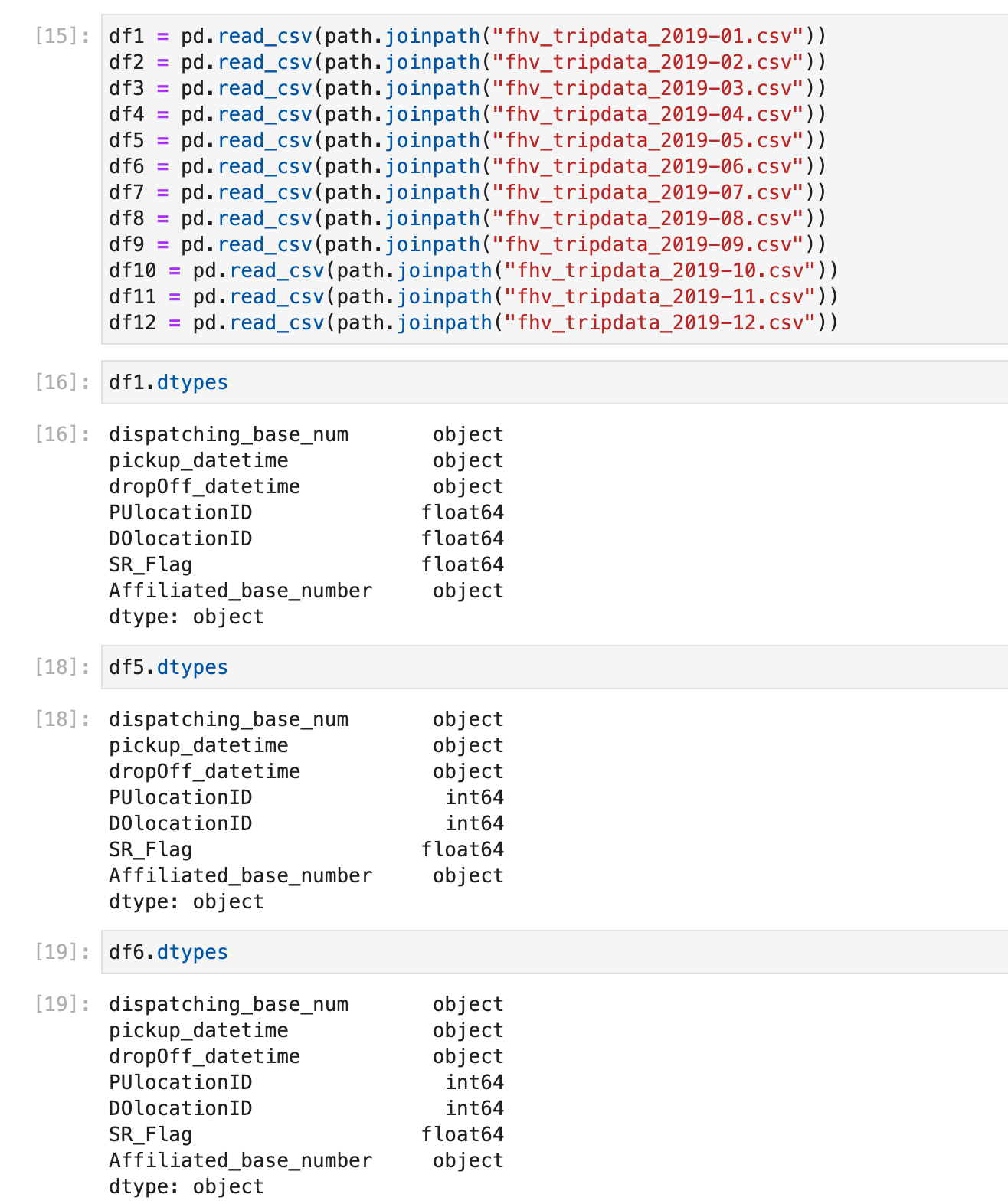
To prevent errors like this, enforce the data types for the columns on the DataFrame before serializing/uploading them to BigQuery:
pd.read_csv("path_or_url").astype({"col1_name": "datatype", "col2_name": "datatype", ..., "colN_name": "datatype"})
# GCS Bucket: Fix Error when importing FHV data to GCS
If you receive the error
gzip.BadGzipFile: Not a gzipped file (b'\n\n')
this is because you have specified the wrong URL to the FHV dataset. Make sure to use:
https://github.com/DataTalksClub/nyc-tlc-data/releases/download/fhv/{dataset_file}.csv.gz
Emphasize the /releases/download part of the URL.
# GCS Bucket - Load Data From URL list in to GCP Bucket
# GCS Bucket - I query my dataset and get a Bad character (ASCII 0) error?
Check the Schema: Ensure that the schema of your dataset is correctly defined.
Formatting Issues: You might have incorrect formatting in your files.
Upload Method: Try uploading the CSV.GZ files without formatting or processing them through pandas. Use
wgetto download if necessary.
# GCP BQ: "bq: command not found"
Run the following command to check if "BigQuery Command Line Tool" is installed or not:
gcloud components list
You can also use bq.cmd instead of bq to make it work.
# GCP: Caution in using BigQuery - bigquery:no
Use BigQuery carefully:
- I created my BigQuery dataset on an account where my free trial was exhausted and received a bill of $80.
- Use BigQuery under free credits and destroy all the datasets after creation.
- Check your billing daily, especially if you've spun up a VM.
# GCP BQ - Cannot read and write in different locations: source: EU, destination: US - Loading data from GCS into BigQuery (different Region):
Be careful when you create your resources on GCP; all of them must share the same region to load data from a GCS Bucket to BigQuery. If you forgot this step, you can create a new dataset in BigQuery using the same region as your GCS Bucket.
This error indicates that your GCS Bucket and the BigQuery dataset are placed in different regions. You need to create a new dataset in BigQuery in the same region as your GCS Bucket and store the data in this newly created dataset.
# GCP BQ - Cannot read and write in different locations: source: <REGION_HERE>, destination: <ANOTHER_REGION_HERE>
Make sure to create the BigQuery dataset in the same location as your GCS Bucket. For instance, if your GCS Bucket was created in us-central1, then the BigQuery dataset must also be created in us-central1.
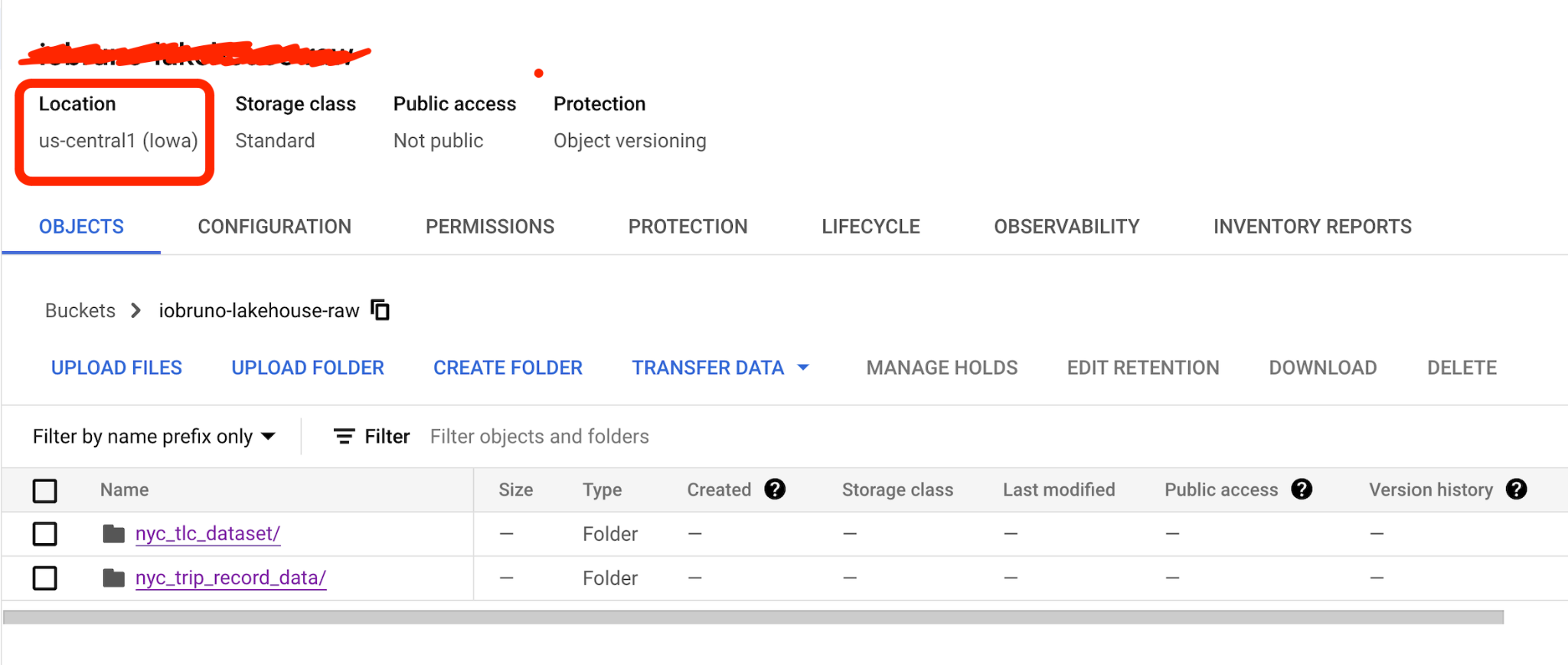
# GCP BQ: Remember to save your queries
It's important to save your progress in the BigQuery SQL Editor frequently.
Here are some tips:
Save Regularly: Use the save button at the top bar in the BigQuery SQL Editor. Your saved queries will be available on the left panel.
Alternative Method: Copy and paste your queries into a file using a text editor like Notepad++ or VS Code. Save it with a
.sqlextension to benefit from syntax highlighting.
By following these methods, you can avoid losing your work in case of unexpected browser issues.
# GCP BQ: Can I use BigQuery for real-time analytics in this project?
While real-time analytics might not be explicitly mentioned, BigQuery has real-time data streaming capabilities, allowing for potential integration in future project iterations.
# GCP BQ: Unable to load data from external tables into a materialized table in BigQuery due to an invalid timestamp error that are added while appending data to the file in Google Cloud Storage
could not parse 'pickup_datetime' as timestamp for field pickup_datetime (position 2)
This error is caused by invalid data in the timestamp column. To resolve this issue:
- Define the schema of the external table using the
STRINGdatatype for the timestamp column. This allows queries to execute without errors. - Filter out the invalid timestamp rows during data import.
- Insert the filtered rows into the materialized table, specifying the
TIMESTAMPdatatype for the timestamp fields.
# GCP BQ: Error Message in BigQuery: annotated as a valid Timestamp, please annotate it as TimestampType(MICROS) or TimestampType(MILLIS)
When you encounter this BigQuery error, it typically relates to how timestamps are stored in Parquet files.
Solution:
To resolve this issue, you can modify the Parquet writing configuration by adding use_deprecated_int96_timestamps=True to the pq.write_to_dataset function. This setting writes timestamps in the INT96 format, which can be more compatible with BigQuery.
Here’s how you can adjust the function:
pq.write_to_dataset(
table,
root_path=root_path,
filesystem=gcs,
use_deprecated_int96_timestamps=True # Write timestamps to INT96 Parquet format
)
References
- Stack Overflow - Parquet compatibility with PyArrow vs PySpark
- Stack Overflow - Editing Parquet files and datetime format errors
- Reddit - Parquet Timestamp to BQ issues
Use the above configuration to ensure compatibility with Google BigQuery when dealing with timestamps in Parquet files.
# GCP BQ - Datetime columns in Parquet files created from Pandas show up as integer columns in BigQuery
Solution:
If you’re using Mage, in the last Data Exporter that writes to Google Cloud Storage, use PyArrow to generate the Parquet file with the correct logical type for the datetime columns. Otherwise, they won't be converted to a timestamp when loaded by BigQuery later on.
import pyarrow as pa
import pyarrow.parquet as pq
import os
if 'data_exporter' not in globals():
from mage_ai.data_preparation.decorators import data_exporter
# Replace with the location of your service account key JSON file.
os.environ['GOOGLE_APPLICATION_CREDENTIALS'] = '/home/src/personal-gcp.json'
bucket_name = "<YOUR_BUCKET_NAME>"
object_key = 'nyc_taxi_data_2022.parquet'
where = f'{bucket_name}/{object_key}'
@data_exporter
def export_data(data, *args, **kwargs):
table = pa.Table.from_pandas(data, preserve_index=False)
gcs = pa.fs.GcsFileSystem()
pq.write_table(
table,
where,
# Convert integer columns in Epoch milliseconds
# to Timestamp columns in microseconds ('us') so
# they can be loaded into BigQuery with the right
# data type
coerce_timestamps='us',
filesystem=gcs
)
Solution 2:
If you’re using Mage, in the last Data Exporter that writes to Google Cloud Storage, provide PyArrow with an explicit schema to generate the Parquet file with the correct logical type for the datetime columns.
schema = pa.schema([
('vendor_id', pa.int64()),
('lpep_pickup_datetime', pa.timestamp('ns')),
('lpep_dropoff_datetime', pa.timestamp('ns')),
('store_and_fwd_flag', pa.string()),
('ratecode_id', pa.int64()),
('pu_location_id', pa.int64()),
('do_location_id', pa.int64()),
('passenger_count', pa.int64()),
('trip_distance', pa.float64()),
('fare_amount', pa.float64()),
('extra', pa.float64()),
('mta_tax', pa.float64()),
('tip_amount', pa.float64()),
('tolls_amount', pa.float64()),
('improvement_surcharge', pa.float64()),
('total_amount', pa.float64()),
('payment_type', pa.int64()),
('trip_type', pa.int64()),
('congestion_surcharge', pa.float64()),
('lpep_pickup_month', pa.int64())
])
table = pa.Table.from_pandas(data, schema=schema)
# GCP: BQ - Create External Table using Python
Reference:
https://cloud.google.com/bigquery/docs/external-data-cloud-storage
Solution:
from google.cloud import bigquery
# Set table_id to the ID of the table to create
table_id = f"{project_id}.{dataset_name}.{table_name}"
# Construct a BigQuery client object
client = bigquery.Client()
# Set the external source format of your table
external_source_format = "PARQUET"
# Set the source_uris to point to your data in Google Cloud
source_uris = [ f'gs://{bucket_name}/{object_key}/*']
# Create ExternalConfig object with external source format
external_config = bigquery.ExternalConfig(external_source_format)
# Set source_uris that point to your data in Google Cloud
external_config.source_uris = source_uris
external_config.autodetect = True
table = bigquery.Table(table_id)
# Set the external data configuration of the table
table.external_data_configuration = external_config
table = client.create_table(table) # Make an API request.
print(f'Created table with external source: {table_id}')
print(f'Format: {table.external_data_configuration.source_format}')
# GCP BQ: Check BigQuery Table Exist And Delete
Reference
Stack Overflow - BigQuery Overwrite Table
Solution
To check if a BigQuery table exists and possibly delete it, utilize the following Python function before using client.create_table:
from google.cloud import bigquery
# Initialize client
client = bigquery.Client()
def table_exists(table_id, client):
"""
Check if a table already exists using the tableID.
:param table_id: str, the ID of the table
:param client: bigquery.Client instance
:return: Boolean
"""
try:
client.get_table(table_id)
return True
except Exception as e: # NotFound:
return False
Use this function to check table existence before creating a new table or taking further actions.
# GCP BQ - Error: Missing close double quote (") character
To avoid this error, you can upload data from Google Cloud Storage to BigQuery through BigQuery Cloud Shell using the command:
bq load --autodetect --allow_quoted_newlines --source_format=CSV dataset_name.table_name "gs://dtc-data-lake-bucketname/fhv/fhv_tripdata_2019-*.csv.gz"
# GCP BQ - Cannot read and write in different locations: source: asia-south2, destination: US
Solution: This problem arises if your GCS and BigQuery storage are in different regions.
One potential way to solve it:
Go to your Google Cloud bucket and check the region in the field named "Location."
In BigQuery, click on the three-dot icon near your project name and select "Create dataset."
In the region field, choose the same region as your Google Cloud bucket.
# GCP BQ - Tip: Using Cloud Function to read csv.gz files from github directly to BigQuery in Google Cloud:
There are multiple benefits of using Cloud Functions to automate tasks in Google Cloud.
Use the following Cloud Function Python script to load files directly into BigQuery. Set your project ID, dataset ID, and table ID accordingly.
import tempfile
import requests
import logging
from google.cloud import bigquery
def hello_world(request):
# table_id = <project_id.dataset_id.table_id>
table_id = 'de-zoomcap-project.dezoomcamp.fhv-2019'
# Create a new BigQuery client
client = bigquery.Client()
for month in range(4, 13):
# Define the schema for the data in the CSV.gz files
url = 'https://github.com/DataTalksClub/nyc-tlc-data/releases/download/fhv/fhv_tripdata_2019-{:02d}.csv.gz'.format(month)
# Download the CSV.gz file from Github
response = requests.get(url)
# Create new table if loading first month data else append
write_disposition_string = "WRITE_APPEND" if month > 1 else "WRITE_TRUNCATE"
# Defining LoadJobConfig with schema of table to prevent it from changing with every table
job_config = bigquery.LoadJobConfig(
schema=[
bigquery.SchemaField("dispatching_base_num", "STRING"),
bigquery.SchemaField("pickup_datetime", "TIMESTAMP"),
bigquery.SchemaField("dropOff_datetime", "TIMESTAMP"),
bigquery.SchemaField("PUlocationID", "STRING"),
bigquery.SchemaField("DOlocationID", "STRING"),
bigquery.SchemaField("SR_Flag", "STRING"),
bigquery.SchemaField("Affiliated_base_number", "STRING"),
],
skip_leading_rows=1,
write_disposition=write_disposition_string,
autodetect=True,
source_format="CSV",
)
# Load the data into BigQuery
# Create a temporary file to prevent the exception: AttributeError: 'bytes' object has no attribute 'tell'
with tempfile.NamedTemporaryFile() as f:
f.write(response.content)
f.seek(0)
job = client.load_table_from_file(
f,
table_id,
location="US",
job_config=job_config,
)
job.result()
logging.info("Data for month %d successfully loaded into table %s.", month, table_id)
return 'Data loaded into table {}.'.format(table_id)
# GCP BQ: When querying two different tables, external and materialized, why do you get the same result with count(distinct(*))?
You need to uncheck cache preferences in query settings

# GCP BQ: How to handle type error from BigQuery and Parquet data?
When injecting data into GCS using Pandas, some datasets might have missing values in the DOlocationID and PUlocationID columns. By default, Pandas will cast these columns as float, leading to inconsistent data types between the Parquet files in GCS and the schema defined in BigQuery. You might encounter the following error:
error: Error while reading table: trips_data_all.external_fhv_tripdata, error message: Parquet column 'DOlocationID' has type INT64 which does not match the target cpp_type DOUBLE.
Solution:
Fix the data type issue in the data pipeline:
Before injecting data into GCS, use
astypeandInt64(which is different fromint64and accepts both missing values and integers) to cast the columns.Example:
df["PUlocationID"] = df.PUlocationID.astype("Int64") df["DOlocationID"] = df.DOlocationID.astype("Int64")It is best to define the data type of all the columns in the Transformation section of the ETL pipeline before loading to BigQuery.
# GCP BQ: Invalid project ID. Project IDs must contain 6-63 lowercase letters, digits, or dashes.
The problem occurs when there is a misplacement of content after the FROM clause in BigQuery SQLs. Check to remove any extra spaces or symbols; ensure project IDs are in lowercase, digits, and dashes only.
# GCP BQ: Does BigQuery support multiple columns partition?
No. Based on the documentation for BigQuery, it does not support more than one column to be partitioned.
# GCP BQ: DATE() Error in BigQuery
Error Message:
PARTITION BY expression must be DATE(<timestamp_column>), DATE(<datetime_column>), DATETIME_TRUNC(<datetime_column>, DAY/HOUR/MONTH/YEAR), a DATE column, TIMESTAMP_TRUNC(<timestamp_column>, DAY/HOUR/MONTH/YEAR), DATE_TRUNC(<date_column>, MONTH/YEAR), or RANGE_BUCKET(<int64_column>, GENERATE_ARRAY(<int64_value>, <int64_value>[, <int64_value>]))
Solution:
Convert the column to datetime first:
# Convert pickup_datetime to datetime
df["pickup_datetime"] = pd.to_datetime(df["pickup_datetime"])
# Convert dropOff_datetime to datetime
df["dropOff_datetime"] = pd.to_datetime(df["dropOff_datetime"])
# GCP BQ: When trying to cluster by DATE(tpep_pickup_datetime) it gives an error: Entries in the CLUSTER BY clause must be column names
No need to convert as you can cluster by a TIMESTAMP column directly in BigQuery. BigQuery supports clustering on TIMESTAMP, DATE, DATETIME, STRING, INT64, and BOOL types.
Clustering sorts data based on the timestamp to optimize queries with filters like WHERE tpep_pickup_datetime BETWEEN ..., rather than creating discrete partitions.
If your goal is to improve performance for time-based queries, combining partitioning by DATE(event_time) and clustering by tpep_pickup_datetime is a good approach.
# GCP BQ - Native tables vs External tables in BigQuery?
Native tables are tables where the data is stored directly in BigQuery. In contrast, external tables store the data outside BigQuery, with BigQuery storing metadata about that external table.
- External tables: These tables are not stored directly in BigQuery but are pulled in from a data lake such as Google Cloud Storage or AWS S3.
- Materialized table: This is a copy of an external table with data stored in BigQuery, consuming storage space.
Resources:
# Why does my partitioned table in BigQuery show as non-partitioned even though BigQuery says it's partitioned?
If your partitioned table in BigQuery shows as non-partitioned, it may be due to a delay in updating the table's details in the UI. The table is likely partitioned, but it may not show the updated information immediately.
Here’s what you can do:
Refresh your BigQuery UI: If you're already inspecting the table in the BigQuery UI, try refreshing the page after a few minutes to ensure the table details are updated correctly.
Open a new tab: Alternatively, try opening a new tab in BigQuery and inspect the table details again. This can sometimes help to load the most up-to-date information.
Be patient: In some cases, there might be a slight delay in reflecting changes, but the table is very likely partitioned.
# GCP BQ ML: Unable to run command (shown in video) to export ML model from BQ to GCS
Issue:
Tried running command to export ML model from BQ to GCS from Week 3:
bq --project_id taxi-rides-ny extract -m nytaxi.tip_model gs://taxi_ml_model/tip_model
It is failing with the following error:
BigQuery error in extract operation: Error processing job Not found: Dataset was not found in location US
I verified the BQ dataset and GCS bucket are in the same region, us-west1. Not sure why it gets location US. I couldn’t find the solution yet.
Solution:
Please ensure the following:
Enter the correct
project_idandgcs_bucketfolder address.Example of a correct GCS bucket folder address:
gs://dtc_data_lake_optimum-airfoil-376815/tip_model
# Dim_zones.sql Dataset was not found in location US When Running fact_trips.sql
To solve this error, specify the location as US when creating the dim_zones table:
{{ config(
materialized='table',
location='US'
) }}
Update this part, re-run the dim_zones creation, and then run fact_trips.
# GCP BQ ML - Export ML model to make predictions does not work for MacBook with Apple M1 chip (arm architecture).
Solution:
Proceed with setting up serving_dir on your computer as described in the extract_model.md file. Then, instead of using:
docker pull tensorflow/serving
use:
docker pull emacski/tensorflow-serving
Then run:
docker run -p 8500:8500 -p 8501:8501 --mount type=bind,source=`pwd`/serving_dir/tip_model,target=/models/tip_model -e MODEL_NAME=tip_model -t emacski/tensorflow-serving
After that, run the curl command as instructed, and you should get a prediction.
Or new since Oct 2024:
Beta release of Docker VMM - the more performant alternative to Apple Virtualization Framework on macOS (requires Apple Silicon and macOS 12.5 or later). https://docs.docker.com/desktop/features/vmm/
# VMs: What do I do if my VM runs out of space?
- Try deleting data you’ve saved to your VM locally during ETLs.
- Kill processes related to deleted files.
- Download
ncduand look for large files (pay particular attention to files related to Prefect). - If you delete any files related to Prefect, eliminate caching from your flow code.
# GCP BQ - External and regular table
External Table
- Data remains stored in a Google Cloud Storage (GCS) bucket.
Regular Table
- Data is copied into BigQuery storage.
Example of creating an external table:
CREATE OR REPLACE EXTERNAL TABLE `your_project.your_dataset.tablenamel`
OPTIONS (
format = 'PARQUET',
uris = ['gs://your-bucket-name/yellow_tripdata_2024-*.parquet']
);
Example of creating a regular table from an external table:
CREATE OR REPLACE TABLE `your_project.your_dataset.tablename`
AS
SELECT * FROM `your_project.your_dataset.yellow_taxi_external`;
Directly loading data from GCS into a regular BigQuery table without creating an external table:
CREATE OR REPLACE TABLE `your_project.your_dataset.yellow_taxi_table`
OPTIONS (
format = 'PARQUET'
) AS
SELECT * FROM `your_project.your_dataset.external_table_placeholder`
FROM EXTERNAL_QUERY(
'your_project.region-us.gcs_external',
'SELECT * FROM `gs://your-bucket-name/yellow_tripdata_2024-*.parquet`'
);
# Can BigQuery work with parquet files directly?
Yes, you can load your Parquet files directly into your GCP (Google Cloud Platform) Bucket first. Then, via BigQuery, you can create an external table of these Parquet files with a query statement like this:
CREATE OR REPLACE EXTERNAL TABLE `module-3-data-warehouse.taxi_data.external_yellow_tripdata_2024`
OPTIONS (
format = 'PARQUET',
uris = ['gs://module3-dez/yellow_tripdata_2024-*.parquet']
);
Make sure to adjust the SQL statement to your own situation and directories. The * symbol can be used as a wildcard to target Parquet files from all the months of 2024.
# Homework: What does it mean “Stop with loading the files into a bucket.”?
What they mean is that they don't want you to do anything more than that. You should load the files into the bucket and create an external table based on those files, but nothing like cleaning the data and putting it in parquet format.
# Homework: Reading parquets from nyc.gov directly into pandas returns Out of bounds error
If you try to read parquets directly from nyc.gov’s cloudfront into pandas, you might encounter this error:
pyarrow.lib.ArrowInvalid: Casting from timestamp[us] to timestamp[ns] would result in out of bounds
Cause:
There is a data record where dropOff_datetime is set to the year 3019 instead of 2019.
Pandas uses "timestamp[ns]" and int64 only allows a ~580-year range, centered on 2000. See pd.Timestamp.max and pd.Timestamp.min.
This becomes out of bounds when pandas tries to read it because 3019 > 2300 (approx value of pd.Timestamp.max).
Fix:
Use pyarrow to read the data:
import pyarrow.parquet as pq df = pq.read_table('fhv_tripdata_2019-02.parquet').to_pandas(safe=False)This will result in unusual timestamps for the offending record.
Read datetime columns separately:
table = pq.read_table('taxi.parquet') datetimes = ['list of datetime column names'] df_dts = pd.DataFrame() for col in datetimes: df_dts[col] = pd.to_datetime(table.column(col), errors='coerce')The
errors='coerce'parameter will convert out-of-bounds timestamps into either the max or min.Remove the offending rows using filter:
import pyarrow.compute as pc table = pq.read_table('taxi.parquet') df = table.filter( pc.less_equal(table["dropOff_datetime"], pa.scalar(pd.Timestamp.max)) ).to_pandas()
# Homework: Uploading files to GCS via GUI
This can help avoid schema issues in the homework. Download files locally and use the 'upload files' button in GCS at the desired path. You can upload many files at once. You can also choose to upload a folder.
# Homework - Qn 5: The partitioned/clustered table isn’t giving me the prediction I expected
Take a careful look at the format of the dates in the question.
# Homework: Qn 6: Did anyone get an exact match for one of the options given in Module 3 homework Q6?
Many people aren’t getting an exact match, but are very close to one of the options. It is suggested to choose the closest option.
# Python: invalid start byte Error Message
UnicodeDecodeError: 'utf-8' codec can't decode byte 0xa0 in position 41721: invalid start byte
Solution:
When reading the data from the web into the pandas dataframe, specify the encoding:
pd.read_csv(dataset_url, low_memory=False, encoding='latin1')When writing the dataframe from the local system to GCS as a CSV, specify the encoding:
df.to_csv(path_on_gsc, compression="gzip", encoding='utf-8')
Alternative: Use pd.read_parquet(url).
# Python: Generators in Python
A generator is a function in Python that returns an iterator using the yield keyword.
A generator is a special type of iterable, similar to a list or a tuple, but with a crucial difference. Instead of creating and storing all the values in memory at once, a generator generates values on-the-fly as you iterate over it. This makes generators memory-efficient, particularly when dealing with large datasets.
# Python: Easiest way to read multiple files at the same time?
The read_parquet function supports a list of files as an argument. The list of files will be merged into a single result table.
# Python: These won't work. You need to make sure you use Int64.
Incorrect:
df['DOlocationID'] = pd.to_numeric(df['DOlocationID'], downcast=integer)
or
df['DOlocationID'] = df['DOlocationID'].astype(int)
Correct:
df['DOlocationID'] = df['DOlocationID'].astype('Int64')
Module 4: Analytics Engineering with dbt
# Warning when run load_yellow_data python script
Warning Details:
RuntimeWarning: As the c extension couldn't be imported, google-crc32c is using a pure python implementation that is significantly slower. If possible, please configure a c build environment and compile extension warnings.warn(_SLOW_CRC32C_WARNING, RuntimeWarning)
Failed to upload ./yellow_tripdata_2024-01.parquet to GCS: Timeout of 120.0s exceeded, last exception: ('Connection aborted.', timeout('The write operation timed out'))
Failed to upload ./yellow_tripdata_2024-03.parquet to GCS: Timeout of 120.0s exceeded, last exception: ('Connection aborted.', timeout('The write operation timed out'))
Issues:
- google-crc32c Warning: The Google Cloud Storage library is using a slow Python implementation instead of the optimized C version.
- Upload Timeout Error: Your file uploads are timing out after 120 seconds.
Solutions:
Install the C-optimized google-crc32c
pip install --upgrade google-crc32cFix Google Cloud Storage Upload Timeout
Solution 1: Increase Timeout
blob.upload_from_filename(file_path, timeout=300) # Set timeout to 5 minutes
# dbt cloud Developer
Please be aware that the demos are done using dbt cloud Developer licensing. Although Team license is available to you upon creation of dbt cloud account for 14 days, the interface won't fully match the demo-ed experience.
# DBT-Config ERROR on CLOUD IDE: No dbt_project.yml found at expected path
Error Details
No dbt_project.yml found at expected path /usr/src/develop/user-70471823426120/environment-70471823422561/repository-70471823410839/dbt_project.yml
Solution Steps
Verify Packages:
- Confirm that every entry in
packages.yml(and their transitive dependencies) includes adbt_project.ymlfile.
- Confirm that every entry in
Initialize Project:
- Use the UI to initialize a new project.
Import Git Repo:
For importing a Git repository of an existing dbt project, follow the instructions available at:
# DBT Cloud production error: prod dataset not available in location EU
Problem:
I am trying to deploy my DBT models to production using DBT Cloud. The data should reside in BigQuery with the dataset location as EU. However, when running the model in production, a prod dataset is incorrectly created in BigQuery with a location of US. This leads to the build failing with the error:
ERROR 404: project.dataset:prod not available in location EU
I have attempted various fixes, but I'm unsure if there is a simpler solution than creating my projects or buckets in the US location.
Note: Everything functions properly in development mode; the issue arises only during job scheduling and execution in production.
Solution:
- Manually create the
proddataset in BigQuery with the EU location specified. - Rerun the production job.
# How do I solve the Dbt Cloud error: prod was not found in location?
You might encounter this error when trying to run dbt in production after following the instructions in the video ‘DE Zoomcamp 4.4.1 - Deployment Using dbt Cloud (Alternative A’):
Database Error in model stg_yellow_tripdata (models/staging/stg_yellow_tripdata.sql)
Not found: Dataset module-4-analytics-eng:prod was not found in location europe-west10
This error is easily resolved. Here are two solutions to address this issue:
Solution #1: Matching the dataset's data location with the source dataset
- Set your ‘prod’ dataset's data location to match the data location of your ‘trips_data_all’ dataset in BigQuery. The dbt process works for the instructor because her ‘prod’ dataset is in the same region as her source data. Since your ‘trips_data_all’ is in europe-west10 (or another region besides US), your prod needs to be there too, not US (which is the default setting when dbt creates a dataset for you in BigQuery).
Solution #2: Changing the dataset to <development dataset>
- Go to: Deploy / Environments / Production (your production environment) / Settings.
- Look at the Deployment credentials. There is an input field called Dataset. The input of ‘prod’ is likely here.
- Replace ‘prod’ with the name of the Dataset that you worked with during development (before moving to Production). This is the Dataset name inside your BigQuery where you successfully ran ‘dbt debug’ and ‘dbt build’.
- After saving, you are ready to rerun your Job!
# Setup: No development environment
Error:
This project does not have a development environment configured. Please create a development environment and configure your development credentials to use the dbt IDE.
The error message provides guidance on resolving this issue. Follow the guide in the dbt cloud setup documentation. Additional instructions can be found in the video @1:42.
# Setup: Connecting dbt Cloud with BigQuery Error
Runtime Error
dbt was unable to connect to the specified database.
The database returned the following error:
Database Error
Access Denied: Project <project_name>: User does not have bigquery.jobs.create permission in project <project_name>.
Check your database credentials and try again. For more information, visit:
Steps to resolve error in Google Cloud:
- Navigate to IAM & Admin and select IAM.
- Click Grant Access if your newly created dbt service account isn't listed.
- In the New principals field, add your service account.
- Select a Role and search for BigQuery Job User to add.
- Go back to dbt Cloud project setup and test your connection.
- Note: Also add BigQuery Data Owner, Storage Object Admin, & Storage Admin to prevent permission issues later in the course.
<{IMAGE:image_id}>
# Setup: Failed to clone repository.
Error: Failed to clone repository.
$ git clone git@github.com:DataTalksClub/data-engineering-zoomcamp.git /usr/src/develop/...
Cloning into '/usr/src/develop/...
Warning: Permanently added '[github.com](http://github.com/),140.82.114.4' (ECDSA) to the list of known hosts.
git@github.com: Permission denied (publickey).
fatal: Could not read from remote repository.
Issue: You don’t have permissions to write to DataTalksClub/data-engineering-zoomcamp.git
Solutions:
Clone the Forked Repository
Clone the repository using your forked repo, which contains your GitHub username. Then, specify the path as:
[your github username]/data-engineering-zoomcamp.gitCreate a Fresh Repo for dbt-lessons
Create a new repository for dbt-lessons. This approach is beneficial as it allows for branching and pull requests without affecting your other repositories. There's no need to create a subfolder for the dbt project files.
Use HTTPS Link
Switch to using an HTTPS link for cloning the repository if SSH access is not configured.
# Errors when I start the server in dbt cloud: Failed to start server. Permission denied (publickey)
Failed to start server. Permission denied (publickey). fatal: Could not read from remote repository. Please make sure you have the correct access rights and the repository exists.
Use the deploy keys in dbt repo details to create a public key in your repo, the issue will be solved.
Steps in detail:
Find dbt Cloud’s SSH Key
- In dbt Cloud, go to Settings > Account Settings > SSH Keys
- Copy the public SSH key displayed there.
Add It to GitHub
- Go to GitHub > Settings > SSH and GPG Keys
- Click "New SSH Key", name it "dbt Cloud", and paste the key.
- Click "Add SSH Key".
Try Restarting dbt Cloud
# dbt: Job - Triggered by pull requests is disabled prerequisites when I try to create a new Continuous Integration job in dbt cloud.
Solution:
Check if you’re on the Developer Plan. As per the prerequisites, you'll need to be enrolled in the Team Plan or Enterprise Plan to set up a CI Job in dbt Cloud.
- If you're on the Developer Plan, you'll need to upgrade to utilize CI Jobs.
Note: A user mentioned that while on the Team Plan (trial period), the option was still disabled. In this case, you might try other troubleshooting steps for the Developer (free) plan.
# Setup: Your IDE session was unable to start. Please contact support.
Issue: If the DBT cloud IDE is loading indefinitely and then giving you this error.
Solution:
- Check the
dbt_cloud_setup.mdfile. - Create an SSH Key.
- Use
git cloneto import the repo into the dbt project. - Copy and paste the deploy key back in your repo settings.
# DBT: I am having problems with columns datatype while running DBT/BigQuery
Issue: If you don’t define the column format while converting from CSV to Parquet, Python will "choose" based on the first rows.
Solution: Define the schema while running the web_to_gcp.py pipeline.
Sebastian adapted the script:
To make the file work with gz files, add the following lines:
- Ensure deletion of the file at the end of each iteration to avoid disk space issues:
file_name_gz = f"{service}_tripdata_{year}-{month}.csv.gz"
open(file_name_gz, 'wb').write(r.content)
os.system(f"gzip -d {file_name_gz}")
os.system(f"rm {file_name_init}.*")
# Parquet: “Parquet column 'ehail_fee' has type DOUBLE which does not match the target cpp_type INT64”
Reason: Parquet files have their own schema. Some Parquet files for green data have records with decimals in the ehail_fee column.
There are some possible fixes:
Drop
ehail_feecolumnDrop the
ehail_feecolumn, as it is not used. For instance, when creating a partitioned table from the external table in BigQuery:SELECT * EXCEPT (ehail_fee) FROM...Modify SQL model
Modify
stg_green_tripdata.sqlmodel with this line:CAST(0 AS NUMERIC) AS ehail_feeModify Airflow DAG
Modify the Airflow DAG to make the conversion and avoid the error:
pv.read_csv(src_file, convert_options=pv.ConvertOptions(column_types={'ehail_fee': 'float64'}))Using Pandas
Fix using Pandas when importing the file from CSV into a DataFrame:
pd.from_csv(..., dtype=type_dict)Note: Regular
int64in Pandas (from the numpy library) does not accept null values (NaN). Use PandasInt64instead. Thetype_dictis a Python dictionary mapping column names to dtypes.
Sources:
# Ingestion: When attempting to use the provided quick script to load trip data into GCS, you receive error Access Denied from the S3 bucket
If the provided URL isn’t working for you (nyc-tlc.s3.amazonaws.com/):
We can use the GitHub CLI to easily download the needed trip data from GitHub and manually upload to a GCS bucket.
Instructions on how to download the CLI here: GitHub
Commands to use:
gh auth login
gh release list -R DataTalksClub/nyc-tlc-data
gh release download yellow -R DataTalksClub/nyc-tlc-data
gh release download green -R DataTalksClub/nyc-tlc-data
Now you can upload the files to a GCS bucket using the GUI.
# Hack to load yellow and green trip data for 2019 and 2020
I initially followed this script but it was taking too long for the yellow trip data. When I downloaded and uploaded the Parquet files directly to GCS, it worked, but there was a schema inconsistency issue when creating the BigQuery table.
I found another solution shared on YouTube, which was suggested by Victoria. You can watch it here:
[Optional] Hack for loading data to BigQuery for Week 4 - YouTube
Make sure to watch until the end, as there are some required schema changes.
# GCP VM: All of sudden ssh stopped working for my VM after my last restart
One common cause experienced is lack of space after running Prefect several times. When running Prefect, check the folder .prefect/storage and delete the logs now and then to avoid the problem.
# GCP FREE TRIAL ACCOUNT ERROR
If you're encountering an error when trying to create a GCP free trial account, and it's not related to country restrictions, credit/debit card problems, or IP issues, it might be a random problem. Here’s a workaround:
- Ask friends in your country to try signing up for the free trial using their Gmail accounts and their debit/credit cards.
- If one succeeds, you can temporarily use their Gmail to access the trial.
This method could help you bypass the issue!
# GCP VM: If you have lost SSH access to your machine due to lack of space. Permission denied (publickey)
You can try to do these steps:

# DBT: When running your first dbt model, if it fails with an error:
404 Not found: Dataset was not found in location US
404 Not found: Dataset eighth-zenith-372015:trip_data_all was not found in location us-west1
To resolve this issue, follow these steps:
Verify Locations in BigQuery:
- Go to BigQuery and check the location of both:
- The source dataset (e.g.,
trips_data_all). - The schema you’re writing to. The name should be in the format
dbt_<first initial><last name>if you didn’t change the default settings.
- The source dataset (e.g.,
- Go to BigQuery and check the location of both:
Check Region Consistency:
- Ensure both datasets are in the same region. Typically, your source data is in your region, while the write location could be multi-regional (e.g., US).
- If there's a mismatch, delete the datasets and recreate them in the specified region with the correct naming format.
Specify Single-Region Location:
- Instead of using a multi-regional location like
US, specify the exact region (e.g.,US-east1). Refer to this Github comment for more details. - Additional guidance is available in this post.
- Instead of using a multi-regional location like
Update Location in DBT Cloud:
- Go to your profile page (top right drop-down --> profile).
- Under Credentials --> Analytics (or your customized name), click on BigQuery >.
- Hit Edit and update your location. You may need to re-upload your service account JSON to refresh your private key. Ensure the region matches exactly as specified in BigQuery.
Following these steps should help resolve location-related errors when running your dbt models.
# DBT: When executing dbt run after installing dbt-utils latest version i.e., 1.0.0 warning has generated
Error: dbt_utils.surrogate_key has been replaced by dbt_utils.generate_surrogate_key
Fix:
- Replace
dbt_utils.surrogate_keywithdbt_utils.generate_surrogate_keyinstg_green_tripdata.sql.
# When executing dbt run after fact_trips.sql has been created, the task failed with error: "Access Denied: BigQuery BigQuery: Permission denied while globbing file pattern."
Fixed by adding the Storage Object Viewer role to the service account in use in BigQuery.
Add the related roles to the service account in use in GCS.
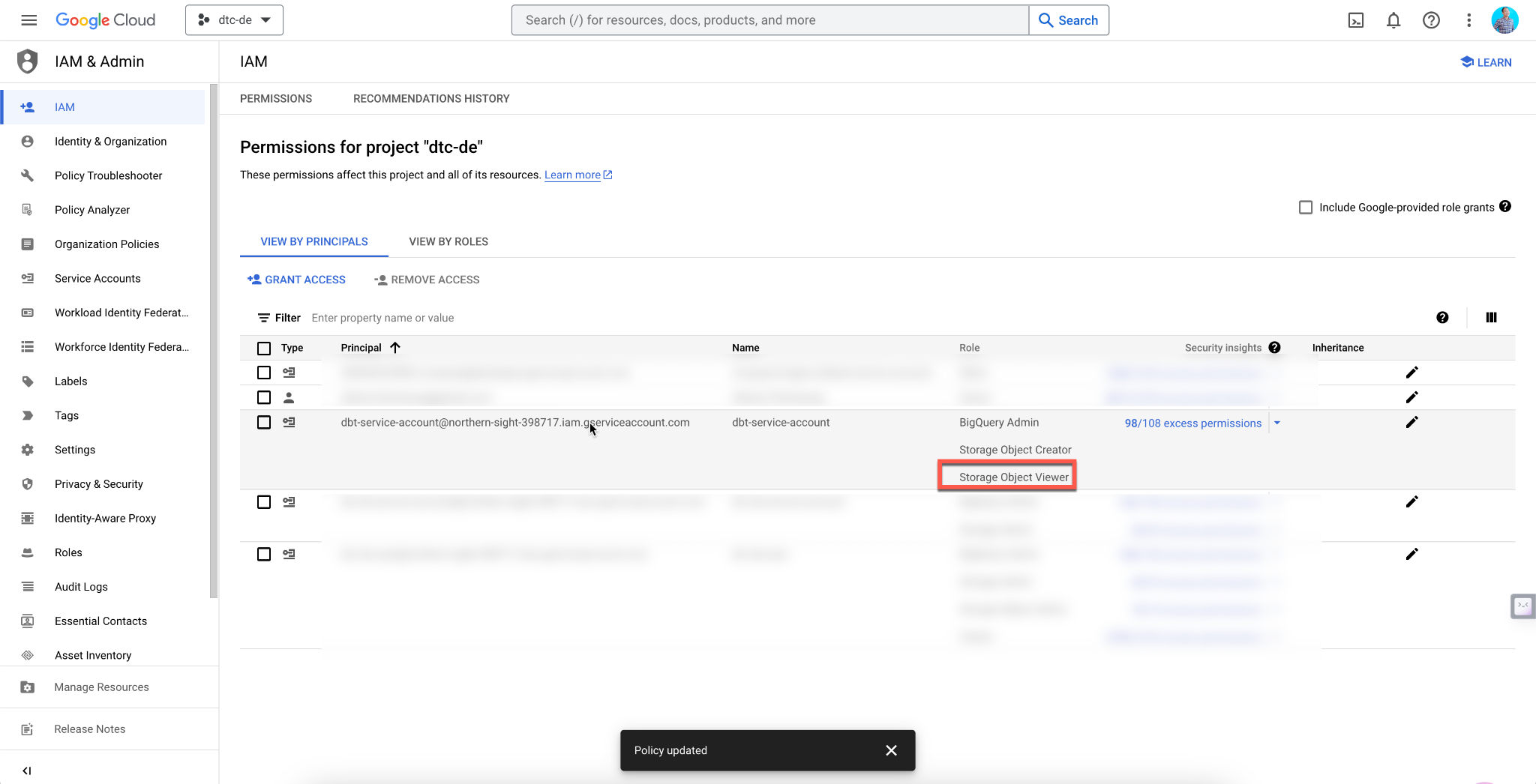
# When you are getting error dbt_utils not found
To resolve the "dbt_utils not found" error, follow these steps:
Create a
packages.ymlfile in the main project directory and add the package metadata:packages: - package: dbt-labs/dbt_utils version: 0.8.0Run the following command:
dbt depsPress Enter.
# Lineage is currently unavailable. Check that your project does not contain compilation errors or contact support if this error persists.
Ensure you properly format your YAML file. Check the build logs if the run was completed successfully. You can expand the command history console (where you type the --vars '{"is_test_run": "false"}') and click on any stage’s logs to expand and read error messages or warnings.
# Build: Why do my Fact_trips only contain a few days of data?
Make sure you use:
dbt run --var 'is_test_run: false'
or
dbt build --var 'is_test_run: false'
Watch out for formatted text from this document: re-type the single quotes. If that does not work, use:
--vars '{"is_test_run": "false"}'
with each phrase separately quoted.
# Build: Why do my fact_trips only contain one month of data?
Check if you specified the if_exists argument correctly when writing data from GCS to BigQuery.
When I wrote my automated flow for each month of the years 2019 and 2020 for green and yellow data, I had specified if_exists="replace" while experimenting with the flow setup. Once you want to run the flow for all months in 2019 and 2020, make sure to set if_exists="append".
if_exists="replace"will replace the whole table with only the month data that you are writing into BigQuery in that one iteration -> you end up with only one month in BigQuery (the last one you inserted).if_exists="append"will append the new monthly data -> you end up with data from all months.
# BigQuery returns an error when I try to run the dm_monthly_zone_revenue.sql model.
After the second SELECT, change this line:
date_trunc('month', pickup_datetime) as revenue_month,
To this line:
date_trunc(pickup_datetime, month) as revenue_month,
Make sure that "month" isn’t surrounded by quotes!
# Replace: {{ dbt_utils.surrogate_key([ field_a, field_b, field_c, …, field_z ]) }}
For this use:
{{ dbt_utils.generate_surrogate_key([ field_a, field_b, field_c, …, field_z ]) }}
Additionally, add a global variable in dbt_project.yml. (...)
# I changed location in dbt, but dbt run still gives me an error
- Remove the dataset from BigQuery that was created by dbt.
- Run
dbt runagain so that it will recreate the dataset in BigQuery with the correct location.
# DBT: I ran dbt run without specifying variable which gave me a table of 100 rows. I ran again with the variable value specified but my table still has 100 rows in BQ.
Remove the dataset from BigQuery created by dbt and run again (with test disabled) to ensure the dataset created has all the rows.
DBT: Why am I getting a new dataset after running my CI/CD Job? / What is this new dbt dataset in BigQuery?
When you create the CI/CD job, under 'Compare Changes against an environment (Deferral)' make sure that you select 'No; do not defer to another environment'. Otherwise, dbt won’t merge your dev models into production models; it will create a new environment called ‘dbt_cloud_pr_number of pull request’
# Why do we need the Staging dataset?
Staging, as the name suggests, acts as an intermediary between raw datasets and the final fact and dimension tables. It helps in transforming raw data into a more usable format. In staging, datasets are typically materialized as views rather than tables.
In the project, you focus on creating production and dbt_name + trips_data_all; the staging dataset serves its role behind the scenes.
# DBT: Docs Served but Not Accessible via Browser
Try removing the network: host line in docker-compose.
# BigQuery adapter: 404 Not found: Dataset was not found in location europe-west6
Go to Account settings > Project > Analytics.
Click on your connection.
Scroll down to Location and type in the GCP location exactly as displayed in GCP (e.g.,
europe-west6). You might need to reupload your GCP key.Delete your dataset in Google BigQuery (GBQ).
Rebuild the project using the command:
dbt buildThe newly built dataset should be in the correct location.
# Dbt+git - Main branch is “read-only”
Create a new branch to edit. More on this can be found here in the dbt docs.
# Dbt+git: It appears that I can't edit the files because I'm in read-only mode. Does anyone know how I can change that?
Create a New Branch:
- You need to create a new branch for development to make changes.
git checkout -b your-feature-branchSwitch to the New Branch:
- This allows you to make edits.
git checkout your-feature-branchCommit and Push Changes:
- Once you've made your changes, commit and push them to the main branch.
git add . git commit -m "Your commit message" git push origin your-feature-branchMerge Changes to Main:
- Finally, merge your changes from your feature branch to the main branch.
# Dbt deploy + Git CI: cannot create CI checks job for deployment to Production
Error:
Triggered by pull requests
This feature is only available for dbt repositories connected through dbt Cloud's native integration with Github, Gitlab, or Azure DevOps
Solution:
Contrary to the guide on DTC repo, don’t use the Git Clone option. Use the Github one instead. A step-by-step guide to unlink Git Clone and relink with Github is available in the next entry.
# Dbt deploy + Git CI - Unable to configure Continuous Integration (CI) with Github
If you’re trying to configure CI with Github and on the job’s options you can’t see Run on Pull Requests? on triggers, follow these steps to reconnect using a native connection instead of cloning by SSH:
Connect Your Github Account
- Go to
Profile Settings > Linked Accounts. - Connect your Github account with the dbt project, allowing the requested permissions.
- More information can be found here.
- Go to
Disconnect Current Configuration
- Navigate to
Account Settings > Projects (analytics) > Github connection. - Click the
Disconnectbutton at the bottom left.
- Navigate to
Reconfigure Github Connection
- After disconnecting, configure again by choosing Github.
- Select your repository from all allowed repositories to work with dbt. Your setup will now be ready.
Configure Triggers
- Go to
Deploy > Job Configuration. - Scroll down to
Triggerswhere you can see the optionRun on Pull Requests:
- Go to
# Compilation Error: Model 'model.my_new_project.stg_green_tripdata' (models/staging/stg_green_tripdata.sql) depends on a source named 'staging.green_trip_external' which was not found
If you're following video DE Zoomcamp 4.3.1 - Building the First DBT Models, you may encounter an issue at 14:25 where the Lineage graph isn't displayed and a Compilation Error occurs, as shown in the attached image.
Don't worry—a quick fix for this is to simply save your schema.yml file. Once you've done this, you should be able to view your Lineage graph without any further issues.
# Compilation Error in test accepted_values_stg_green_tripdata_Payment_type__False___var_payment_type_values_ (models/staging/schema.yml) 'NoneType' object is not iterable
In the macro test_accepted_values (found in tests/generic/builtin.sql), an error was triggered by the test accepted_values_stg_green_tripdata_Payment_type__False___var_payment_type_values_ located in models/staging/schema.yml.
To resolve this issue, ensure the following variable is added to your dbt_project.yml file:
vars:
payment_type_values: [1, 2, 3, 4, 5, 6]
# dbt: macro errors with get_payment_type_description(payment_type)
You will face this issue if you copied and pasted the exact macro directly from the data-engineering-zoomcamp repo.
Error Message
BigQuery adapter: Retry attempt 1 of 1 after error: BadRequest('No matching signature for operator CASE for argument types: STRING, INT64, STRING, INT64, STRING, INT64, STRING, INT64, STRING, INT64, STRING, INT64, STRING, NULL at [35:5]; reason: invalidQuery, location: query, message: No matching signature for operator CASE for argument types: STRING, INT64, STRING, INT64, STRING, INT64, STRING, INT64, STRING, INT64, STRING, INT64, STRING, NULL at [35:5]')
Solution
To resolve this issue, change the data type of the numbers (1, 2, 3, etc.) to text by enclosing them in quotes. The payment_type data type should be a string.
Updated Macro
{#
This macro returns the description of the payment_type
#}
{% macro get_payment_type_description(payment_type) -%}
case {{ payment_type }}
when '1' then 'Credit card'
when '2' then 'Cash'
when '3' then 'No charge'
when '4' then 'Dispute'
when '5' then 'Unknown'
when '6' then 'Voided trip'
end
{%- endmacro %}
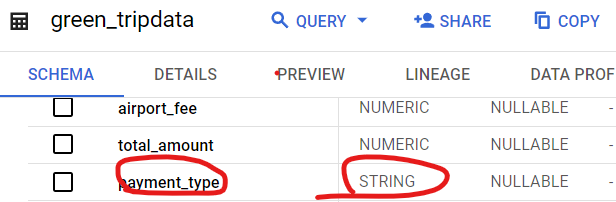
# Troubleshooting in dbt:
The dbt error log contains a link to BigQuery. When you follow it, you will see your query, and the problematic line will be highlighted.
# DBT: Why changing the target schema to “marts” actually creates a schema named “dbt_marts” instead?
It is a default behavior of dbt to append custom schema to the initial schema. To override this behavior, create a macro named generate_schema_name.sql:
{% macro generate_schema_name(custom_schema_name, node) -%}
{%- set default_schema = target.schema -%}
{%- if custom_schema_name is none -%}
{{ default_schema }}
{%- else -%}
{{ custom_schema_name | trim }}
{%- endif -%}
{%- endmacro %}
Now you can override the default custom schema in dbt_project.yml.
# How to set subdirectory of the github repository as the dbt project root
There is a project setting which allows you to set Project subdirectory in dbt cloud:
# Compilation Error: Model 'model.XXX' (models/<model_path>/XXX.sql) depends on a source named '<a table name>' which was not found
Remember to modify your .sql models to read from existing table names in BigQuery/Postgres DB.
Example:
select * from {{ source('staging', '<your table name in the database>') }}
# Compilation Error: Model '<model_name>' (<model_path>) depends on a node named '<seed_name>' which was not found (Production Environment)
Make sure that you create a pull request from your Development branch to the Production branch (main by default). After that, check in your seeds folder to ensure the seed file is inside it. Another thing to check is your .gitignore file. Make sure that the .csv extension is not included.
# When executing dbt run after using fhv_tripdata as an external table: you get "Access Denied: BigQuery BigQuery: Permission denied"
Go to your dbt cloud service account.
Add the
Storage Object AdminandStorage Adminroles in addition toBigQuery Admin.
# How to automatically infer the column data type (pandas missing value issues)?
Problem: When injecting data to BigQuery, you may face a type error. This is because pandas by default will parse integer columns with missing values as float type.
Solution:
One way to solve this problem is to specify or cast the data type as Int64 during the data transformation stage.
If specifying all the integer columns is inconvenient, you can use convert_dtypes to infer the data type automatically.
- Make pandas infer the correct data type (as pandas parse int with missing as float):
# Fill missing values with a placeholder
df.fillna(-999999, inplace=True)
# Infer data types
df = df.convert_dtypes()
# Replace placeholder with None
df = df.replace(-999999, None)
# When loading GitHub repo raise exception that 'taxi_zone_lookup' not found
Seed files are loaded from a directory named 'seed'. Therefore, you should rename the directory currently named 'data' to 'seed'.
# ‘taxi_zone_lookup’ not found
Check the .gitignore file and make sure you don’t have *.csv in it.
If you're encountering a dbt error with the following message:
Runtime Error in rpc request (from remote system.sql)
404 Not found: Table dtc-de-0315:trips_data_all.green_tripdata_partitioned was not found in location europe-west6
Location: europe-west6
Job ID: 168ee9bd-07cd-4ca4-9ee0-4f6b0f33897c
- Verify that all your datasets are configured with the correct region. For example, use
europe-west6instead of a general region likeEU. - To update the region in dbt settings:
- Go to
dbt -> projects -> optional settings - Manually set the location to match the required region.
- Go to
# Data type errors when ingesting with parquet files
The easiest way to avoid these errors is by ingesting the relevant data in a .csv.gz file type. Then, do:
CREATE OR REPLACE EXTERNAL TABLE `dtc-de.trips_data_all.fhv_tripdata`
OPTIONS (
format = 'CSV',
uris = ['gs://dtc_data_lake_dtc-de-updated/data/fhv_all/fhv_tripdata_2019-*.csv.gz']
);
This example should help you avoid data type issues for week 4.
# Inconsistent number of rows when re-running fact_trips model
This issue arises from the way deduplication is handled in two staging files.
Solution:
- Add an
ORDER BYclause in thePARTITION BYsection of both staging files. - Continue adding columns to the
ORDER BYclause until the row count in thefact_tripstable is consistent upon re-running the model.
Explanation:
We partition by vendor_id and pickup_datetime, selecting the first row (rn=1) from these partitions. These partitions lack an order, so every execution might yield a different first row. The inconsistency leads to different rows being processed, possibly with or without an unknown borough. Consequently, the fact_trips model discards a varying number of rows based on the presence of unknown boroughs.
# Data Type Error when running fact table
If you encounter a data type error on the trip_type column, it may be due to some nan values that aren't null in BigQuery.
Solution: Try casting it to FLOAT datatype instead of NUMERIC.
SELECT CAST(trip_type AS FLOAT) FROM your_table;
# CREATE TABLE has columns with duplicate name locationid.
This error could result if you are using a SELECT * query without specifying the table names.
Example:
WITH dim_zones AS (
SELECT * FROM `engaged-cosine-374921`.`dbt_victoria_mola`.`dim_zones`
WHERE borough != 'Unknown'
),
fhv AS (
SELECT * FROM `engaged-cosine-374921`.`dbt_victoria_mola`.`stg_fhv_tripdata`
)
SELECT * FROM fhv
INNER JOIN dim_zones AS pickup_zone
ON fhv.PUlocationID = pickup_zone.locationid
INNER JOIN dim_zones AS dropoff_zone
ON fhv.DOlocationID = dropoff_zone.locationid;
To resolve, replace with:
SELECT fhv.* FROM fhv
# Bad int64 value: 0.0 error
Some ehail fees are null, causing a Bad int64 value: 0.0 error when casting them to integer.
Solution:
Use
safe_cast, which returns NULL instead of throwing an error. Implementsafe_castfrom thedbt_utilsfunction in Jinja code for casting into integer as follows:{{ dbt_utils.safe_cast('ehail_fee', api.Column.translate_type("integer")) }} as ehail_fee,Alternatively, use
safe_cast(ehail_fee as integer)without relying ondbt_utils.
# Bad int64 value: 2.0/1.0 error
You might encounter this when building the fact_trips.sql model. The issue may be with the payment_type_description field.
Using safe_cast as above would cause the entire field to become null. A better approach is to drop the offending decimal place, then cast to integer:
cast(replace({{ payment_type }},'.0','') as integer)
# Bad int64 value: 1.0 error (again)
I found that there are more columns causing the bad INT64: ratecodeid and trip_type on Green_tripdata table. You can use the queries below to address them:
CAST(
REGEXP_REPLACE(CAST(rate_code AS STRING), r'\.0', '') AS INT64
) AS ratecodeid,
CAST(
CASE
WHEN REGEXP_CONTAINS(CAST(trip_type AS STRING), r'\.\d+') THEN NULL
ELSE CAST(trip_type AS INT64)
END AS INT64
) AS trip_type
# DBT: Error on building fact_trips.sql - Parquet column 'ehail_fee' has type DOUBLE which does not match the target cpp_type INT64.
To resolve the error regarding the 'ehail_fee' column type mismatch, you can use the following line in stg_green_trips.sql to replace the original ehail_fee line:
{{ dbt.safe_cast('ehail_fee', api.Column.translate_type("numeric")) }} as ehail_fee,
This ensures that the column type is correctly converted to match the expected type.
# The - vars argument must be a YAML dictionary, but was of type str
Remember to add a space between the variable and the value. Otherwise, it won't be interpreted as a dictionary.
Correct usage:
dbt run --var 'is_test_run: false'
Not able to change Environment Type as it is greyed out and inaccessible. You don't need to change the environment type. If you are following the videos, you are creating a Production Deployment, so the only available option is the correct one.
# Access Denied: Table yellow_tripdata: User does not have permission to query table yellow_tripdata, or perhaps it does not exist in location US.
Error Details
Database Error in model stg_yellow_tripdata (models/staging/stg_yellow_tripdata.sql)
Access Denied: Table taxi-rides-ny-339813-412521:trips_data_all.yellow_tripdata: User does not have permission to query table taxi-rides-ny-339813-412521:trips_data_all.yellow_tripdata, or perhaps it does not exist in location US.
compiled Code at target/run/taxi_rides_ny/models/staging/stg_yellow_tripdata.sql
Solution
Branch Verification:
- Ensure you are working on the correct branch. If not, switch to the appropriate branch.
Schema Configuration:
Edit the
04-analytics-engineering/taxi_rides_ny/models/staging/schema.ymlfile.Ensure the configuration is correct:
sources: - name: staging database: your_database_name
Custom Branch Setup in dbt Cloud:
If the error persists, consider running the dbt job on a custom branch:
- Navigate to the environment settings in dbt Cloud.
- In General settings, select "Only run on a custom branch".
- Enter the name of your custom branch (e.g., HW).
- Click Save.
# Could not parse the dbt project. Please check that the repository contains a valid dbt project.
Running the Environment on the master branch causes this error. You must activate the “Only run on a custom branch” checkbox and specify the branch you are working on when the Environment is set up.
# Made change to your modeling files and commit to your development branch, but Job still runs on old file?
Switch to the main branch and make a pull request from the development branch. This will take you to GitHub. Approve the merge and rerun your job; it should work as planned now.
# Setup: I’ve set Github and Bigquery to dbt successfully. Why nothing showed in my Develop tab?
Before you can develop some data model on dbt, you should:
Create a Development Environment: Ensure that your development environment is properly configured.
Set Parameters: Specify necessary parameters within the environment.
Once the model has been developed, also create a deployment environment to create and run jobs.
# BigQuery returns an error when I try to run ‘dbt run’:
My taxi data was loaded into GCS with etl_web_to_gcs.py, which converts CSV data into Parquet. Then I placed raw data trips into external tables, and when I executed dbt run, I got an error message:
Parquet column 'passenger_count' has type INT64 which does not match the target cpp_type DOUBLE.
This is because several columns in the files have different data formats.
To resolve this error, I added the transformation:
df[col] = df[col].astype('Int64')
Apply this transformation to the following columns:
passenger_countpayment_typeRatecodeIDVendorIDtrip_type
After making these changes, the process completed successfully.
# DBT: Running `dbt run --models stg_green_tripdata --var 'is_test_run: false'` is not returning anything:
Use the syntax below instead if the code in the tutorial is not working.
dbt run --select stg_green_tripdata --vars '{"is_test_run": false}'
# DBT: Error: No module named 'pytz' while setting up dbt with docker
Following dbt with BigQuery on Docker readme.md, after running docker-compose build and docker-compose run dbt-bq-dtc init, you might encounter the error:
ModuleNotFoundError: No module named 'pytz'
Solution:
Add the following line in the Dockerfile under FROM --platform=$build_for python:3.9.9-slim-bullseye as base:
RUN python -m pip install --no-cache pytz
# VS Code: NoPermissions (FileSystemError): Error: EACCES: permission denied (linux)
If you encounter problems editing dbt_project.yml when using Docker after running docker-compose run dbt-bq-dtc init, to change the profile ‘taxi_rides_ny’ to 'bq-dbt-workshop’, execute the following command:
sudo chown -R username path
If you see the error "DBT - Internal Error: Profile should not be None if loading is completed" when running dbt debug, change the directory to the newly created subdirectory, such as the taxi_rides_ny directory, which contains the dbt project.
# Google Cloud BigQuery: Location Problems
When running a query on BigQuery, you might encounter the error: "This table is not on the specified location".
To resolve this issue, consider the following steps:
Check the Locations: Ensure the locations of your bucket, datasets, and tables are consistent. They should all reside in the same location.
Modify Query Settings:
- Go to the query window.
- Select More -> Query Settings.
- Select the correct location.
Verify Table Paths: Double-check the paths in your query:
- Click on the table.
- Go to Details.
- Copy the correct path.
# DBT Deploy: This dbt Cloud run was cancelled because a valid dbt project was not found.
This issue occurs when the dbt project is moved to another directory in the repository or if you're on a different branch than expected.
Solution:
Navigate to the projects window on dbt Cloud.
Go to Settings -> Edit.
Add the directory path where the dbt project is located. Ensure that this path matches your file explorer path. For example:
/week5/taxi_rides_nyCheck that there are no files waiting to be committed to GitHub if you’re running the job to deploy to PROD.
- Ensure the PROD environment is set up to check the main branch, or the specified branch.
In the image below, the branch "ella2024" is set to be checked as "production-ready" by the "freshness" check mark in PROD environment settings. Each time a branch is merged into "ella2024" and a PR is triggered, the CI check job initiates. Note that merging and closing the PR must be done manually.
- Set up the PROD custom branch (if not the default main) in the Environment setup screen.
# DBT Deploy + CI: Location Problems on BigQuery
When creating a pull request and running the CI, dbt creates a new schema on BigQuery. By default, this new schema is created with the 'US' location. If your datasets, schemas, and tables are in the 'EU', this will cause an error and the pull request will not be accepted.
To change the location to 'EU' in the connection to BigQuery from dbt, follow these steps:
- Navigate to Dbt -> Project -> Settings -> Connection BigQuery
- Open Optional Settings
- Set Location to
EU
# DBT Deploy: Error When trying to run the dbt project on Prod
When running the dbt project on production, ensure the following steps:
Pull Request and Merge
- Make the pull request and merge the branch into the main.
Version Check
- Ensure you have the latest version if changes were made to the repository elsewhere.
Project File Accessibility
- Verify that the
dbt_project.ymlfile is accessible to the project. If not, refer to the solution for the error: "Dbt: This dbt Cloud run was cancelled because a valid dbt project was not found."
- Verify that the
Dataset Consistency
- Confirm that the name assigned to the dataset on BigQuery matches the name specified in the production environment configuration on dbt Cloud.
# DBT: Error: “404 Not found: Dataset <dataset_name>:<dbt_schema_name> was not found in location EU” after building from stg_green_tripdata.sql
In the step in this video (DE Zoomcamp 4.3.1 - Build the First dbt Models), after creating stg_green_tripdata.sql and clicking build, I encountered an error saying the dataset was not found in location EU. The default location for dbt Bigquery is the US, so when generating the new Bigquery schema for dbt, unless specified, the schema locates in the US.
Solution:
- Ensure the location is set to
EUwhen adding connection details:- Navigate to Develop -> Configure Cloud CLI -> Projects -> taxi_rides_ny -> (connection) Bigquery -> Edit
- Under Location (Optional), type
EU - Save the changes.
# Homework: Ingesting FHV_20?? data
Issue
If you’re having problems loading the FHV_20?? data from the GitHub repo into GCS and then into BigQuery (input file not of type parquet), follow these steps:
Append URL Template
Add
?raw=trueto theURL_TEMPLATElink:URL_TEMPLATE = URL_PREFIX + "/fhv_tripdata_{{ execution_date.strftime('%Y-%m') }}.parquet?raw=true"Update URL Prefix
Ensure
URL_PREFIXis set to the following value:URL_PREFIX = "https://github.com/alexeygrigorev/datasets/blob/master/nyc-tlc/fhv"It is critical to use this link with the keyword
blob. If the link containstree, replace it withblob. Everything else, including thecurl -sSLfcommand, can remain unchanged.
# Ingesting FHV: alternative with kestra
Add this task based on the previous ones:
id: if_fhv_taxi
type: io.kestra.plugin.core.flow.If
condition: "{{inputs.taxi == 'fhv'}}"
then:
id: bq_fhv_tripdata
type: io.kestra.plugin.gcp.bigquery.Query
sql: |
CREATE TABLE IF NOT EXISTS `{{kv('GCP_PROJECT_ID')}}.{{kv('GCP_DATASET')}}.fhv_tripdata` ( unique_row_id BYTES OPTIONS (description = 'A unique identifier for the trip, generated by hashing key trip attributes.'), filename STRING OPTIONS (description = 'The source filename from which the trip data was loaded.'), dispatching_base_num STRING, pickup_datetime TIMESTAMP, dropoff_datetime TIMESTAMP, PUlocationID NUMERIC, DOlocationID NUMERIC, SR_Flag STRING, Affiliated_base_number STRING ) PARTITION BY DATE(pickup_datetime);
id: bq_fhv_table_ext
type: io.kestra.plugin.gcp.bigquery.Query
sql: |
CREATE OR REPLACE EXTERNAL TABLE `{{kv('GCP_PROJECT_ID')}}.{{render(vars.table)}}_ext` ( dispatching_base_num STRING, pickup_datetime TIMESTAMP, dropoff_datetime TIMESTAMP, PUlocationID NUMERIC, DOlocationID NUMERIC, SR_Flag STRING, Affiliated_base_number STRING ) OPTIONS ( format = 'CSV', uris = ['{{render(vars.gcs_file)}}'], skip_leading_rows = 1, ignore_unknown_values = TRUE );id: bq_fhv_table_tmp
type: io.kestra.plugin.gcp.bigquery.Query
sql: |
CREATE OR REPLACE TABLE `{{kv('GCP_PROJECT_ID')}}.{{render(vars.table)}}` AS SELECT MD5(CONCAT( COALESCE(CAST(pickup_datetime AS STRING), ""), COALESCE(CAST(dropoff_datetime AS STRING), ""), COALESCE(CAST(PUlocationID AS STRING), ""), COALESCE(CAST(DOLocationID AS STRING), "") )) AS unique_row_id, "{{render(vars.file)}}" AS filename, * FROM `{{kv('GCP_PROJECT_ID')}}.{{render(vars.table)}}_ext`;id: bq_fhv_merge
type: io.kestra.plugin.gcp.bigquery.Query
sql: |
MERGE INTO `{{kv('GCP_PROJECT_ID')}}.{{kv('GCP_DATASET')}}.fhv_tripdata` T USING `{{kv('GCP_PROJECT_ID')}}.{{render(vars.table)}}` S ON T.unique_row_id = S.unique_row_id WHEN NOT MATCHED THEN INSERT (unique_row_id, filename, dispatching_base_num, pickup_datetime, dropoff_datetime, PUlocationID, DOlocationID, SR_Flag, Affiliated_base_number) VALUES (S.unique_row_id, S.filename, S.dispatching_base_num, S.pickup_datetime, S.dropoff_datetime, S.PUlocationID, S.DOlocationID, S.SR_Flag, S.Affiliated_base_number);
Add a trigger too:
id: fhv_schedule
type: io.kestra.plugin.core.trigger.Schedule
cron: "0 11 1 * *"
inputs:
taxi: fhv
And modify inputs:
inputs:
- id: taxi
type: SELECT
displayName: Select taxi type
values: [yellow, green, fhv]
defaults: green
# Homework: Ingesting NYC TLC Data
The easiest way to upload datasets from GitHub for the homework is by utilizing this script: git_csv_to_gcs.py. It is similar to a script provided in 03-data-warehouse/extras/web_to_gcs.py. <{IMAGE:image_id}>
# How to set environment variable easily for any credentials
If you need to securely set up credentials for a project and intend to push it to a git repository, using an environment variable is a recommended option.
For example, for scripts like web_to_gcs.py or git_csv_to_gcs.py, you may need to set these variables:
GOOGLE_APPLICATION_CREDENTIALSGCP_GCS_BUCKET
The easiest option to manage this is to use a .env file with dotenv.
To install and utilize this package, follow these steps:
Install
python-dotenv:pip install python-dotenvAdd the following code to inject these variables into your project:
from dotenv import load_dotenv import os # Load environment variables from .env file load_dotenv() # Now you can access environment variables like GCP_GCS_BUCKET and GOOGLE_APPLICATION_CREDENTIALS credentials_path = os.getenv("GOOGLE_APPLICATION_CREDENTIALS") BUCKET = os.environ.get("GCP_GCS_BUCKET")
# Invalid date types after Ingesting FHV data through CSV files: Could not parse 'pickup_datetime' as a timestamp
If you uploaded manually the FHV 2019 CSV files, you may face errors regarding date types. Try to create an external table in BigQuery but define the pickup_datetime and dropoff_datetime to be strings:
CREATE OR REPLACE EXTERNAL TABLE `gcp_project.trips_data_all.fhv_tripdata` (
dispatching_base_num STRING,
pickup_datetime STRING,
dropoff_datetime STRING,
PUlocationID STRING,
DOlocationID STRING,
SR_Flag STRING,
Affiliated_base_number STRING
)
OPTIONS (
format = 'csv',
uris = ['gs://bucket/*.csv']
);
Then, when creating the FHV core model in dbt, use TIMESTAMP(CAST(()) to ensure it first parses as a string and then converts it to a timestamp:
WITH fhv_tripdata AS (
SELECT * FROM {{ ref('stg_fhv_tripdata') }}
),
dim_zones AS (
SELECT * FROM {{ ref('dim_zones') }}
WHERE borough != 'Unknown'
)
SELECT fhv_tripdata.dispatching_base_num,
TIMESTAMP(CAST(fhv_tripdata.pickup_datetime AS STRING)) AS pickup_datetime,
TIMESTAMP(CAST(fhv_tripdata.dropoff_datetime AS STRING)) AS dropoff_datetime,
# Invalid data types after Ingesting FHV data through parquet files: Could not parse SR_Flag as Float64, Couldn’t parse datetime column as timestamp, couldn’t handle NULL values in PULocationID, DOLocationID
If you uploaded the FHV 2019 parquet files manually after downloading from this source, you may face errors regarding data types while loading the data into a landing table (e.g., fhv_tripdata). To avoid these errors, create an external table with the schema defined as follows and load each month in a loop:
CREATE OR REPLACE EXTERNAL TABLE `dw-bigquery-week-3.trips_data_all.external_tlc_fhv_trips_2019` (
dispatching_base_num STRING,
pickup_datetime TIMESTAMP,
dropoff_datetime TIMESTAMP,
PUlocationID FLOAT64,
DOlocationID FLOAT64,
SR_Flag FLOAT64,
Affiliated_base_number STRING
)
OPTIONS (
format = 'PARQUET',
uris = ['gs://project id/fhv_2019_8.parquet']
);
You can also use:
uris = ['gs://project id/fhv_2019_*.parquet']
This approach removes the need for a loop and allows you to process all months in a single run.
# Join Error on LocationID: "Unable to find common supertype for templated argument"
No matching signature for operator = for argument types: STRING, INT64
Signature: T1 = T1
Error: Unable to find common supertype for templated argument.
Solution:
Make sure the LocationID field is of the same type in both tables. If it is in string format in one table, use the following dbt code to convert it to an integer:
{{ dbt.safe_cast("PULocationID", api.Column.translate_type("integer")) }} as pickup_locationid
# Google Looker Studio: you have used up your 30-day trial
When accessing Looker Studio through the Google Cloud Project console, you may be prompted to subscribe to the Pro version and receive the following errors:
Instead, navigate to https://lookerstudio.google.com/navigation/reporting which will take you to the free version.
# How does dbt handle dependencies between models?
Dbt provides a mechanism called ref to manage dependencies between models. By referencing other models using the ref keyword in SQL, dbt automatically understands the dependencies and ensures the correct execution order.
# Loading FHV Data goes into slumber using Mage?
Try loading the data using Jupyter notebooks in a local environment. There might be bandwidth issues with Mage.
Load the data into a pandas DataFrame using the URLs, make necessary transformations, upload to the GCP bucket, or alternatively download the Parquet/CSV files locally and then upload to GCP manually.
# Region Mismatch in DBT and BigQuery
If you are using the datasets copied into BigQuery from BigQuery public datasets, the region will be set as US by default. It is much easier to set your dbt profile location as US while transforming the tables and views. You can change the location as follows:
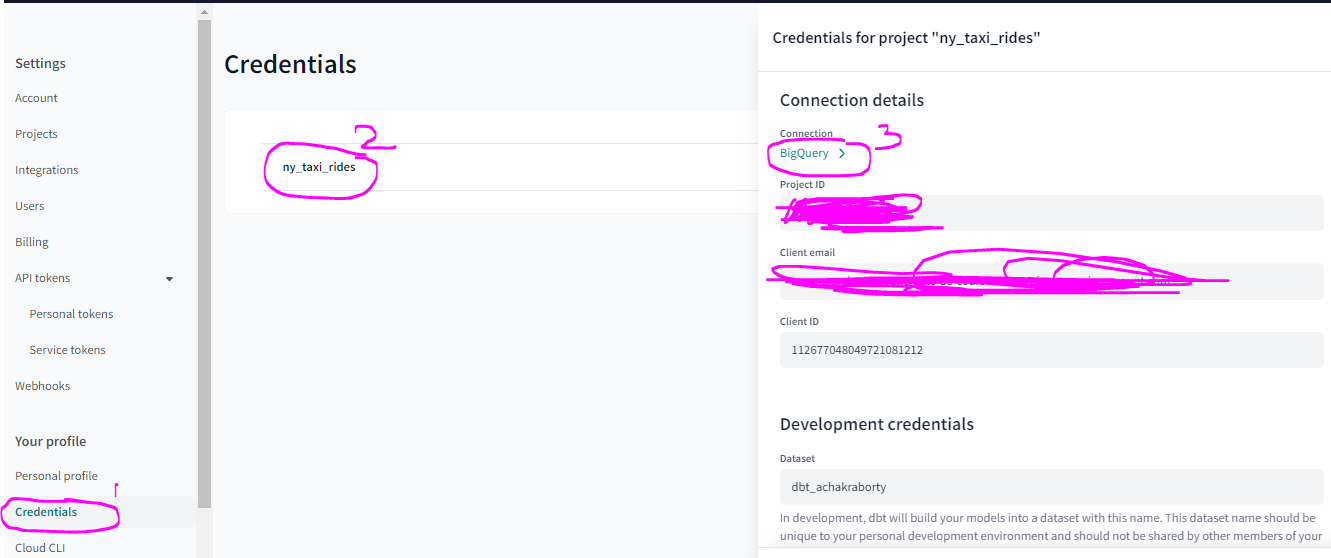
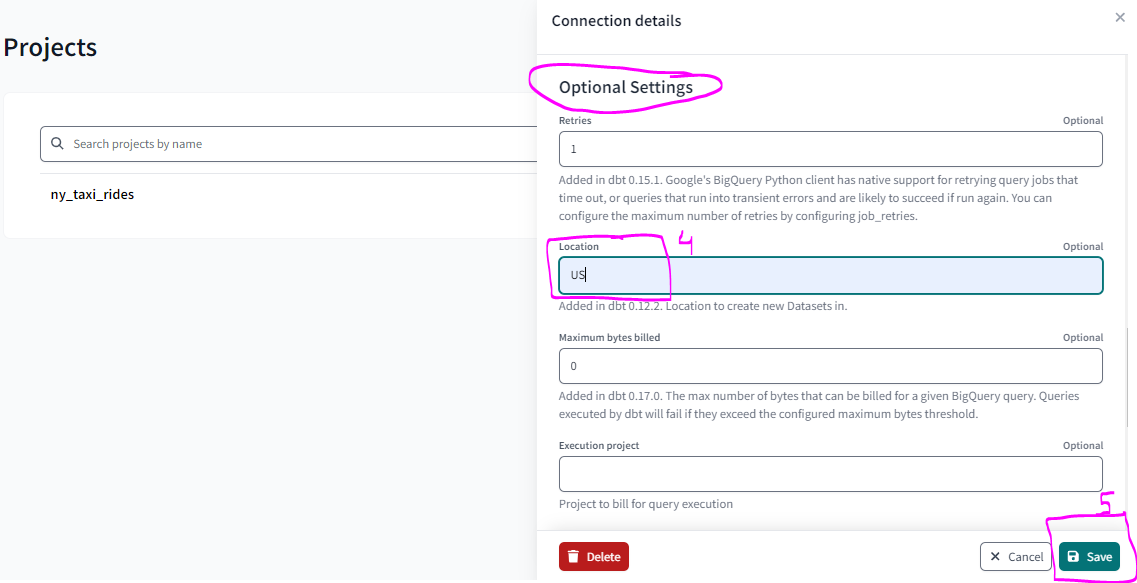
# What is the fastest way to upload taxi data to dbt-postgres?
Use the PostgreSQL COPY FROM feature, which is compatible with CSV files.
Steps:
Create the Table
First, create your table (example):
CREATE TABLE taxis ( … );Use COPY Functionality
Use the
COPYcommand (example):COPY taxis FROM PROGRAM 'url' WITH ( FORMAT csv, HEADER true, ENCODING utf8 );- Syntax for
COPY:
COPY table_name [ ( column_name [, ...] ) ] FROM { 'filename' | PROGRAM 'command' | STDIN } [ [ WITH ] ( option [, ...] ) ] [ WHERE condition ]- Syntax for
# dbt: Where should we create `profiles.yml`?
For local environments using dbt-core, the profile configuration is valid for all projects. Note: dbt Cloud doesn’t require it.
The ~/.dbt/profiles.yml file should be located in your user's home directory. On Windows, this would typically be:
C:\Users\<YourUsername>\.dbt\profiles.yml
Replace <YourUsername> with your actual Windows username. This file is used by dbt to store connection profiles for different projects.
Here's how you can create the profiles.yml file in the appropriate directory:
- Open File Explorer and navigate to
C:\Users\<YourUsername>\. - Create a new folder named
.dbtif it doesn't already exist. - Inside the
.dbtfolder, create a new file namedprofiles.yml.
Usage example can be found here.
# dbt: Are there UI options for dbt Core like dbt Cloud?
While dbt Core does not have an official UI like dbt Cloud, there are several tools available that provide UI functionality:
Altimate's VS Code Extension:
- Use the VS Code dbt Power User extension.
- Sign up for the community plan for free usage at Altimate and add the API into your VS Code extension.
VSCode Snippets for dbt and Jinja:
- Access the snippets package for SQL, YAML, and Markdown here.
Monitoring with Elementary:
- Monitor dbt projects using Elementary.
- Learn more about its setup and features in this Medium article.
# When configuring the profiles.yml file for dbt-postgres with jinja templates with environment variables, I'm getting "Credentials in profile \"PROFILE_NAME\", target: 'dev', invalid: '5432' is not of type 'integer'"
Make sure that the port number is set as an integer in your profiles.yml file. Environment variables are usually strings, so you need to explicitly convert them to integers in Jinja. Update the line that sets the port with something like:
port: {{ env_var('DB_PORT') | int }}
This will ensure that the value is treated as an integer.
# DBT: The database is correct but I get Error with Incorrect Schema in Models
What to do if your dbt model fails with an error similar to:
DBT-CORE
Check profiles.yml:
- Ensure your
profiles.ymlfile is correctly configured with the correct schema and database under your target. This file is typically located in~/.dbt/.
Example configuration:
DBT-CLOUD-IDE
Check Credentials in dbt Cloud UI:
- Navigate to the Credentials section in the dbt Cloud project settings.
- Ensure the correct database and schema are set (e.g., ‘my_dataset’).

Verify Environment Settings:
- Double-check that you are working in the correct environment (dev, prod, etc.), as dbt Cloud allows different settings for different environments.
No Need for profiles.yml:
- In dbt Cloud, you don’t need to configure
profiles.ymlmanually. All connection settings are handled via the UI.
# DBT: DBT allows only 1 project in free developer version.
Yes, DBT allows only 1 project under one account. But you can create multiple accounts as shown below:
Module 5: Spark
# Documentation or book sign not shown even after doing `dbt docs generate`.
In the free version, it does not show the docs when models are run in the development environment. Create a production job and tick the 'generate docs' section. Execute it, and it will generate the documentation.
# Setting up Java and Spark (with PySpark) on Linux (Alternative option using SDKMAN):
Install SDKMAN:
curl -s "https://get.sdkman.io" | bash source "$HOME/.sdkman/bin/sdkman-init.sh"Using SDKMAN, install Java 11 and Spark 3.3.2:
sdk install java 11.0.22-tem sdk install spark 3.3.2Open a new terminal or run the following in the same shell:
source "$HOME/.sdkman/bin/sdkman-init.sh"Verify the locations and versions of Java and Spark that were installed:
echo $JAVA_HOME java -version echo $SPARK_HOME spark-submit --version
# PySpark: Setting Spark up in Google Colab
If you’re struggling to set things up "locally" (meaning non-managed environments like your laptop, a VM, or Codespaces), you can follow this guide to use Spark in Google Colab:
Launch Spark on Google Colab and Connect to SparkUI
Starter notebook:
GitHub Repository - Spark in Colab
It’s advisable to spend some time setting up locally rather than using this solution immediately.
# Spark-shell: unable to load native-hadoop library for platform - Windows
If after installing Java (either JDK or OpenJDK), Hadoop, and Spark, and setting the corresponding environment variables, you encounter the following error when running spark-shell in CMD:
java.lang.IllegalAccessError: class org.apache.spark.storage.StorageUtils$ (in unnamed module @0x3c947bc5) cannot access class sun.nio.ch.DirectBuffer (in module java.base) because module java.base does not export sun.nio.ch to unnamed module @0x3c947bc5
Solution:
- Java 17 or 19 is not supported by Spark.
- Spark 3.x requires Java 8, 11, or 16.
- Install Java 11 from the website provided in the windows.md setup file.
# PySpark: Python was not found; run without arguments to install from the Microsoft Store, or disable this shortcut from Settings > Manage App Execution Aliases.
I encountered this error while executing a user-defined function in Spark (crazy_stuff_udf). I am working on Windows and using conda. After following the setup instructions, I discovered that the PYSPARK_PYTHON environment variable was not set correctly, as conda has different Python paths for each environment.
Solution:
Run the following command inside the appropriate environment:
pip install findsparkAdd the following to the top of your script:
import findspark findspark.init()
# PySpark: TypeError: code() argument 13 must be str, not int, while executing `import pyspark` (Windows/ Spark 3.0.3 - Python 3.11)
This error occurs because Python 3.11 has some inconsistencies with the older Spark 3.0.3 version.
Solutions:
Downgrade Python Version:
- Use Python 3.9. A conda environment can help in managing different Python versions.
Upgrade PySpark Version:
- Install a newer PySpark version, such as 3.5.1 or above, which is compatible with Python 3.11.
# Example commands
conda create -n pyspark_env python=3.9
conda activate pyspark_env
pip install pyspark==3.5.1
Ensure that your environment is set up correctly to avoid version mismatches.
# Import pyspark - Error: No Module named ‘pyspark’
Ensure that your PYTHONPATH is set correctly to include the PySpark library. You can check if PySpark is pointing to the correct location by running:
import pyspark
print(pyspark.__file__)
It should point to the location where PySpark is installed (e.g., /home/<your username>/spark/spark-3.x.x-bin-hadoop3.x/python/pyspark/__init__.py).
# Cannot find Spark jobs UI at localhost
This is because the current port is in use, so the Spark UI will run on a different port. You can check which port Spark is using by running this command:
spark.sparkContext.uiWebUrl
If it indicates a different port, you should access that specific port instead. Additionally, ensure that there are no other notebooks or processes that might be using the same port. Clean up unused resources to avoid port conflicts.
# Java+Spark - Easy setup with miniconda env (worked on MacOS)
If you want to manage all Python dependencies within the same virtual environment (e.g., conda), along with Java and Spark, follow these steps:
Install OpenJDK 11:
On MacOS, run:
brew install java11Add the following line to your
~/.bashrcor~/zshrc:export PATH="/opt/homebrew/opt/openjdk@11/bin:$PATH"Activate Your Environment:
Use your preferred tool (pipenv, poetry, or conda) to activate your working environment.
Install PySpark:
Run:
pip install pysparkProceed with Exercises:
Work with your Spark exercises as usual. All default Spark commands will be available in the shell session under the activated environment.
Note: You don’t need
findsparkfor initialization.Resolving Py4J Errors:
If you encounter an error like:
Py4JJavaError: An error occurred while calling (...) java.net.ConnectException: Connection refused: no further information;Ensure you're using compatible versions of JDK or Python with Spark. Spark 3.5.0 supports JDK 8/11/17 and Python 3.8+.
Use SDKMan! to install:
sdk install java 17.0.10-librca sdk install spark 3.5.0 sdk install hadoop 3.3.5Create and activate a conda environment with:
conda create -n ENV_NAME python=3.11 conda activate ENV_NAME pip install pyspark==3.5.0Windows Py4J Setup:
Ensure correct PATH settings in your
~/.bashrc:export JAVA_HOME="/c/tools/jdk-11.0.21" export PATH="${JAVA_HOME}/bin:${PATH}" export HADOOP_HOME="/c/tools/hadoop-3.2.0" export PATH="${HADOOP_HOME}/bin:${PATH}" export SPARK_HOME="/c/tools/spark-3.3.2-bin-hadoop3" export PATH="${SPARK_HOME}/bin:${PATH}" export PYTHONPATH="${SPARK_HOME}/python/:$PYTHONPATH" export PYTHONPATH="${SPARK_HOME}spark-3.5.1-bin-hadoop3py4j-0.10.9.5-src.zip:$PYTHONPATH"Download
winutilsfrom Stephenlaye2/winutils3.3.0 and place them in/c/tools/hadoop-3.2.0/bin.Check out this video for a solution: How To Resolve Issue with Writing DataFrame to Local File
Restart your IDE and computer to apply the changes. Be aware that fixing one error might result in new ones like
o31.parquet. Address these as needed.
# Spark - Installation Error Code 1603
Issue: Spark installation on Windows completed but failed to run.
This is a common Windows Installer error code indicating that there was a fatal error during installation. It often occurs due to issues like insufficient permissions, conflicts with other software, or problems with the installer package.
Step to Solve the Issue:
Installing Chocolatey
Chocolatey is a package manager for Windows, which makes it easy to install, update, and manage software.
Installation Steps
Open PowerShell as an Administrator
- Press
Win + Xand select Windows PowerShell (Admin) or search for PowerShell, right-click, and select Run as administrator.
- Press
Run the following command to install Chocolatey:
Set-ExecutionPolicy Bypass -Scope Process -Force; \ [System.Net.ServicePointManager]::SecurityProtocol = [System.Net.ServicePointManager]::SecurityProtocol -bor 3072; \ iex ((New-Object System.Net.WebClient).DownloadString('https://community.chocolatey.org/install.ps1'))Verify the installation
- Close and reopen PowerShell as an administrator and run:
choco -v- You should see the Chocolatey version number indicating that it has been installed successfully.
Command for Global Acceptance
- To globally accept all licenses for all packages installed using Chocolatey, run the following command:
choco feature enable -n allowGlobalConfirmation- This command configures Chocolatey to automatically accept license agreements for all packages, streamlining the installation process and avoiding prompts for each package.
# RuntimeError: Java gateway process exited before sending its port number
After installing everything, including PySpark, when running this script in a Jupyter Notebook:
import pyspark
from pyspark.sql import SparkSession
spark = SparkSession.builder \
.master("local[*]") \
.appName('test') \
.getOrCreate()
# Read the CSV file
df = spark.read \
.option("header", "true") \
.csv('taxi+_zone_lookup.csv')
df.show()
You might encounter the error:
RuntimeError: Java gateway process exited before sending its port number
Solutions
Solution 1:
Install
findsparkby running:pip install findsparkAdd the following lines to the top of your script:
import findspark findspark.init()
Solution 2:
Ensure that PySpark points to the correct location. Run:
pyspark.__file__It should list a path like
/home/<your user name>/spark/spark-3.0.3-bin-hadoop3.2/python/pyspark/__init__.pyif you followed the setup correctly.If it points to your Python site-packages, remove the PySpark directory there.
Verify
.bashrcfor correct exports, ensuring no conflicting variables are present.
Alternative Solution:
Set environment variables permanently at the system and user levels by following a tutorial.
Once installed, skip to 7:14 in the tutorial to help set up environment variables.
# Module Not Found Error in Jupyter Notebook.
Even after installing pyspark correctly on a Linux machine (VM) as instructed in the course, a module not found error was encountered in the Jupyter Notebook.
The solution that worked:
Run the following in the Jupyter Notebook:
!pip install findsparkImport and initialize
findspark:import findspark findspark.init()Thereafter, import
pysparkand create the Spark context as usual.
If the above solution does not work, try:
- Using
!pip3 install pysparkinstead of!pip install pyspark.
To filter based on conditions across multiple columns:
from pyspark.sql.functions import col
new_final.filter((new_final.a_zone=="Murray Hill") & (new_final.b_zone=="Midwood")).show()
# Py4JJavaError - ModuleNotFoundError: No module named 'py4j' while executing `import pyspark`
To resolve the ModuleNotFoundError: No module named 'py4j' when executing import pyspark, follow these steps:
Check for Py4J File Version:
- Run the command:
ls ${SPARK_HOME}/python/lib/ - Note the version of the
py4jfile.
- Run the command:
Update the
PYTHONPATH:- Use the version identified above to update the
PYTHONPATHaccordingly. For example:export PYTHONPATH="${SPARK_HOME}/python/lib/py4j-0.10.9.3-src.zip:$PYTHONPATH" - Ensure that the version matches the filename under
${SPARK_HOME}/python/lib/.
- Use the version identified above to update the
Verify Spark's Py4J Version:
- Check the Py4J version of the Spark you're using from the Apache Spark documentation.
Install Py4J (if needed):
- If the above steps do not resolve the issue, run:
pip install py4j - This can resolve version mismatches or missing installations.
- If the above steps do not resolve the issue, run:
This should address the ModuleNotFoundError for Py4J when using PySpark. Ensure all versions are consistent and correct.
# Py4J Error - ModuleNotFoundError: No module named 'py4j' (Solve with latest version)
To resolve the Py4J error, follow these steps:
Install the latest available Py4J version using conda:
conda install -c conda-forge py4jMake sure to replace with the latest version number as found on the website.
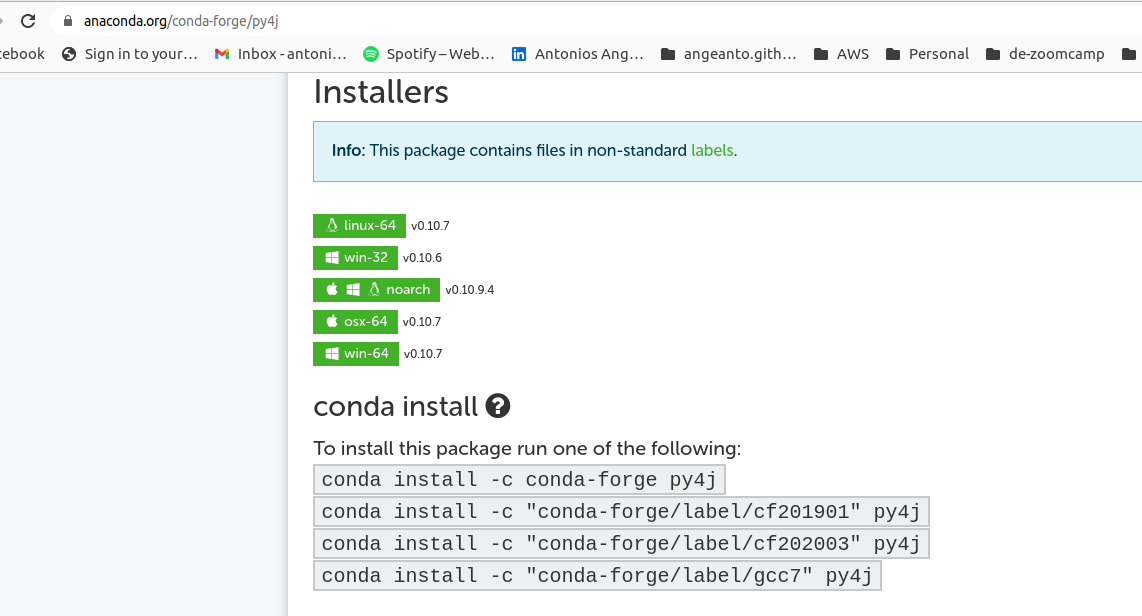
Add the following lines to your
.bashrcfile:export PYTHONPATH="${SPARK_HOME}/python/:$PYTHONPATH" export PYTHONPATH="${SPARK_HOME}/python/lib/py4j-0.10.9.7-src.zip:$PYTHONPATH"
# Exception: Jupyter command `jupyter-notebook` not found.
Even after exporting paths correctly, you may find that Jupyter is installed but jupyter-notebook is not found. Follow these steps to resolve the issue:
Full steps:
Update and Upgrade Packages:
sudo apt update && sudo apt -y upgradeInstall Python:
sudo apt install python3-pip python3-devInstall Python Virtualenv:
sudo -H pip3 install --upgrade pip sudo -H pip3 install virtualenvCreate a Python Virtual Environment:
mkdir notebook cd notebook virtualenv jupyterenv source jupyterenv/bin/activateInstall Jupyter Notebook:
pip install jupyterRun Jupyter Notebook:
jupyter notebook
For full instructions, refer to this guide or the original instructions here.
# Following 5.2.1, I am getting an error - Head:cannot open ‘taxi+_zone_lookup.csv’ for reading: No such file or directory
The latest filename is just taxi_zone_lookup.csv, so it should work after removing the + now.
# Error: java.io.FileNotFoundException
# Code executed:
df = spark.read.parquet(pq_path)
# … some operations on df …
df.write.parquet(pq_path, mode="overwrite")
# Error:
# java.io.FileNotFoundException: File file:/home/xxx/code/data/pq/fhvhv/2021/02/part-00021-523f9ad5-14af-4332-9434-bdcb0831f2b7-c000.snappy.parquet does not exist
The problem is that Spark performs lazy transformations, so the actual action that triggers the job is df.write, which deletes the parquet files it tries to read (mode="overwrite").
Solution:
Write to a different directory:
df.write.parquet(pq_path_temp, mode="overwrite")
# Hadoop: FileNotFoundException: Hadoop bin directory does not exist, when trying to write (Windows)
Create the Hadoop /bin directory manually and add the downloaded files there. The shell script for Windows installation typically places them in /c/tools/hadoop-3.2.0/.
# Which type of SQL is used in Spark? Postgres? MySQL? SQL Server?
Spark uses its own type of SQL, known as Spark SQL.
The SQL syntax across various providers is generally similar, as shown below:
SELECT [attributes]
FROM [table]
WHERE [filter]
GROUP BY [grouping attributes]
HAVING [filtering the groups]
ORDER BY [attribute to order]
(INNER/FULL/LEFT/RIGHT) JOIN [table2]
ON [attributes table joining table2] (...)
What differs most between SQL providers are the built-in functions.
For built-in Spark SQL functions, check this link: Spark SQL Functions
Extra information on Spark SQL:
# The spark viewer on localhost:4040 was not showing the current run
Solution:
Ensure you have identified all running Spark notebooks. If multiple notebooks are running, each will attempt to use available ports starting from 4040.
If a port is in use, Spark automatically assigns the next available port (e.g., 4041, 4042, etc.).
To find the exact port being used by your current Spark application, run the following command:
spark.sparkContext.uiWebUrlThe result will provide the URL, for example:
[172.19.10.61:4041](http://172.19.10.61:4041).If the expected port does not show your current run, verify that cleanup has been performed on closed or non-running containers.
# Java: java.lang.NoSuchMethodError: sun.nio.ch.DirectBuffer.cleaner()Lsun/misc/Cleaner Error during repartition call (conda pyspark installation)
java.lang.NoSuchMethodError: sun.nio.ch.DirectBuffer.cleaner()Lsun/misc/Cleaner;
Solution: Replace Java Developer Kit 11 with Java Developer Kit 8.
# Java - RuntimeError: Java gateway process exited before sending its port number
This error indicates that the JAVA_HOME environment variable is not set correctly in the notebook log.
For more information, you can refer to the following resource:
PySpark Exception - Java gateway process exited before sending the driver its port number
# Spark fails when reading from BigQuery and using `.show()` on `SELECT` queries
I got it working using gcs-connector-hadoop-2.2.5-shaded.jar and Spark 3.1.
I also added the google_credentials.json and .p12 to authenticate with GCS. These files are downloadable from the GCP Service account.
To create the SparkSession:
spark = SparkSession.builder.master('local[*]') \
.appName('spark-read-from-bigquery') \
.config('BigQueryProjectId', 'razor-project-xxxxxxx') \
.config('BigQueryDatasetLocation', 'de_final_data') \
.config('parentProject', 'razor-project-xxxxxxx') \
.config("google.cloud.auth.service.account.enable", "true") \
.config("credentialsFile", "google_credentials.json") \
.config("GcpJsonKeyFile", "google_credentials.json") \
.config("spark.driver.memory", "4g") \
.config("spark.executor.memory", "2g") \
.config("spark.memory.offHeap.enabled", True) \
.config("spark.memory.offHeap.size", "5g") \
.config('google.cloud.auth.service.account.json.keyfile', "google_credentials.json") \
.config("fs.gs.project.id", "razor-project-xxxxxxx") \
.config("fs.gs.impl", "com.google.cloud.hadoop.fs.gcs.GoogleHadoopFileSystem") \
.config("fs.AbstractFileSystem.gs.impl", "com.google.cloud.hadoop.fs.gcs.GoogleHadoopFS") \
.getOrCreate()
# Spark: BigQuery connector Automatic configuration
To automatically configure the Spark BigQuery connector, you can create a SparkSession by specifying the spark.jars.packages configuration.
spark = SparkSession.builder
.master('local')
.appName('bq')
.config("spark.jars.packages", "com.google.cloud.spark:spark-bigquery-with-dependencies_2.12:0.23.2")
.getOrCreate()
This approach automatically downloads the required dependency jars and configures the connector, eliminating the need to manually manage this dependency. More details are available here.
# Spark: How to read from GCP data lake using PySpark?
There are a few steps to read from Google Cloud Storage (GCS) using PySpark:
Download the Cloud Storage connector for Hadoop
- You can download it here.
- This
.jarfile connects PySpark with GCS.
Move the .jar file to your Spark file directory
- If you installed Spark using Homebrew on MacOS, create a
/jarsdirectory under your Spark directory, e.g.,/opt/homebrew/Cellar/apache-spark/3.2.1/jars.
- If you installed Spark using Homebrew on MacOS, create a
Import necessary classes in your Python script
import pyspark from pyspark.sql import SparkSession from pyspark.conf import SparkConf from pyspark.context import SparkContextSet up your configurations before building the SparkSession
conf = SparkConf() \ .setMaster('local[*]') \ .setAppName('test') \ .set("spark.jars", "/opt/homebrew/Cellar/apache-spark/3.2.1/jars/gcs-connector-hadoop3-latest.jar") \ .set("spark.hadoop.google.cloud.auth.service.account.enable", "true") \ .set("spark.hadoop.google.cloud.auth.service.account.json.keyfile", "path/to/google_credentials.json") sc = SparkContext(conf=conf) sc._jsc.hadoopConfiguration().set("fs.AbstractFileSystem.gs.impl", "com.google.cloud.hadoop.fs.gcs.GoogleHadoopFS") sc._jsc.hadoopConfiguration().set("fs.gs.impl", "com.google.cloud.hadoop.fs.gcs.GoogleHadoopFileSystem") sc._jsc.hadoopConfiguration().set("fs.gs.auth.service.account.json.keyfile", "path/to/google_credentials.json") sc._jsc.hadoopConfiguration().set("fs.gs.auth.service.account.enable", "true")Build your SparkSession with the new configurations
spark = SparkSession.builder \ .config(conf=sc.getConf()) \ .getOrCreate()You can now read files directly from GCS!
Note: If you encounter start-slave.sh: command not found, ensure Spark is correctly installed and paths are set.
# How can I read a small number of rows from the parquet file directly?
To read a small number of rows from a parquet file, you can use PyArrow or Apache Spark:
Using PyArrow
from pyarrow.parquet import ParquetFile
pf = ParquetFile('fhvhv_tripdata_2021-01.parquet')
# PyArrow builds tables, not dataframes
tbl_small = next(pf.iter_batches(batch_size=1000))
# Convert the table to a DataFrame
df = tbl_small.to_pandas()
Alternatively, without PyArrow
df = spark.read.parquet('fhvhv_tripdata_2021-01.parquet')
df1 = df.sort('DOLocationID').limit(1000)
pdf = df1.select("*").toPandas()
# DataType error when creating Spark DataFrame with a specified schema?
When defining the schema for a Spark DataFrame, you might encounter a data type error if you're using a Parquet file with the schema definition from the TLC example. The error occurs because the PULocationID and DOLocationID columns are defined as IntegerType, but the Parquet file uses INT64.
You'll get an error like:
Parquet column cannot be converted in file [...] Column [...] Expected: int, Found: INT64
To resolve this issue:
- Change the schema definition from
IntegerTypetoLongType. This adjustment should align the expected and actual data types, allowing the DataFrame to be created successfully.
# Remove white spaces from column names in Pyspark
df_finalx = df_finalw.select([col(x).alias(x.replace(" ", "")) for x in df_finalw.columns])
# AttributeError: 'DataFrame' object has no attribute 'iteritems'
This error occurs in the Spark video 5.3.1 - First Look at Spark/PySpark because the example utilizes CSV files from 2021, but the current data is in parquet format.
When running the command:
spark.createDataFrame(df1_pandas).show()
You may encounter the Attribute error due to incompatibility between pandas version 2.0.0 and Spark 3.3.2. To resolve this, you can:
Downgrade pandas to version 1.5.3 using the following command:
pip install -U pandas==1.5.3Alternatively, add the following line after importing pandas if you prefer not to downgrade:
pd.DataFrame.iteritems = pd.DataFrame.items
This issue is resolved in Spark versions from 3.4.1 onwards.
# AttributeError: 'DataFrame' object has no attribute 'iteritems'
Another alternative is to install pandas 2.0.1 (it worked well at the time of writing this), and it is compatible with Pyspark 3.5.1. Make sure to add or edit your environment variable like this:
export SPARK_HOME="${HOME}/spark/spark-3.5.1-bin-hadoop3"
export PATH="${SPARK_HOME}/bin:${PATH}"
# Spark Standalone Mode on Windows
To set up Spark in standalone mode on Windows, follow these steps:
Open a CMD terminal in administrator mode.
Navigate to your Spark home directory:
cd %SPARK_HOME%Start a master node:
bin\spark-class org.apache.spark.deploy.master.MasterStart a worker node:
bin\spark-class org.apache.spark.deploy.worker.Worker spark://<master_ip>:<port> --host <IP_ADDR>Example:
bin\spark-class org.apache.spark.deploy.worker.Worker spark://localhost:7077 --host <IP_ADDR>spark://<master_ip>:<port>: Copy the address from the previous command. For example,spark://localhost:7077- Use
--host <IP_ADDR>if you want to run the worker on a different machine. You can leave it empty for local setup.
Access the Spark UI through
localhost:8080.
# Export PYTHONPATH command in Linux is temporary
To make the export PYTHONPATH command permanent, consider the following options:
Add to
.bashrc:- Open the
.bashrcfile located in your home directory using a text editor, such asnanoorvim.
nano ~/.bashrc- Add the
export PYTHONPATHline, such as:
export PYTHONPATH="/your/custom/path"- Save the changes and source the file to apply the changes:
source ~/.bashrc- Open the
Run specific commands in Python scripts:
- You can initialize the environment directly in a Python script using:
import findspark findspark.init()
By using these methods, you ensure that the PYTHONPATH is set up for every session automatically.
# Compression Error: zcat output is gibberish, seems like still compressed
In the code along from Video 5.3.3, Alexey downloads the CSV files from the NYT website and gzips them in their bash script. Currently (2023), if we download the data from the GH course repo, it is already zipped as csv.gz files. Following the video exactly would zip them again, leading to gibberish output when using zcat, as it only unzips the file once.
Solution: Do not gzip the files downloaded from the course repo. Simply use wget to download and save them as csv.gz files. Then the zcat command and the showSchema command will work correctly.
URL="${URL_PREFIX}/${TAXI_TYPE}/${TAXI_TYPE}_tripdata_${YEAR}-${FMONTH}.csv.gz"
LOCAL_PREFIX="data/raw/${TAXI_TYPE}/${YEAR}/${FMONTH}"
LOCAL_FILE="${TAXI_TYPE}_tripdata_${YEAR}_${FMONTH}.csv.gz"
LOCAL_PATH="${LOCAL_PREFIX}/${LOCAL_FILE}"
echo "downloading ${URL} to ${LOCAL_PATH}"
mkdir -p ${LOCAL_PREFIX}
wget ${URL} -O ${LOCAL_PATH}
echo "compressing ${LOCAL_PATH}"
# gzip ${LOCAL_PATH} <- uncomment this line
# PicklingError: Could not serialise object: IndexError: tuple index out of range.
This error occurs while running:
spark.createDataFrame(df_pandas).show()
Cause
This issue is usually related to the Python version. As of March 2, 2023, Spark does not support Python 3.11.
Solution
Create a New Environment with a Supported Python Version:
Using Conda, you can create a virtual environment with Python 3.10:
conda create -n myenv python=3.10 anacondaActivate the Environment:
To use Python 3.10, activate the environment:
conda activate myenvDeactivate the Environment:
If you need to return to the initial setup, you can deactivate the environment:
conda deactivate
# Connecting from local Spark to GCS: Spark does not find my Google credentials as shown in the video?
Make sure you have your credentials for your GCP in your VM under the location defined in the script.
# Spark docker-compose setup
To run Spark in a Docker setup:
Build Bitnami Spark Docker
a. Clone the Bitnami repository using the command:
git clone https://github.com/bitnami/containers.git(Tested on commit 9cef8b892d29c04f8a271a644341c8222790c992)
b. Edit the file
bitnami/spark/3.3/debian-11/Dockerfileand update the Java and Spark version as follows:"python-3.10.10-2-linux-${OS_ARCH}-debian-11" "java-17.0.5-8-3-linux-${OS_ARCH}-debian-11"Reference: GitHub
c. Build the Docker image by navigating to the above directory and running the Docker build command:
cd bitnami/spark/3.3/debian-11/ docker build -t spark:3.3-java-17 .Run Docker Compose
Use the following
docker-compose.ymlfile:version: '2' services: spark: image: spark:3.3-java-17 environment: - SPARK_MODE=master - SPARK_RPC_AUTHENTICATION_ENABLED=no - SPARK_RPC_ENCRYPTION_ENABLED=no - SPARK_LOCAL_STORAGE_ENCRYPTION_ENABLED=no - SPARK_SSL_ENABLED=no volumes: - "./:/home/jovyan/work:rw" ports: - '8080:8080' - '7077:7077' spark-worker: image: spark:3.3-java-17 environment: - SPARK_MODE=worker - SPARK_MASTER_URL=spark://spark:7077 - SPARK_WORKER_MEMORY=1G - SPARK_WORKER_CORES=1 - SPARK_RPC_AUTHENTICATION_ENABLED=no - SPARK_RPC_ENCRYPTION_ENABLED=no - SPARK_LOCAL_STORAGE_ENCRYPTION_ENABLED=no - SPARK_SSL_ENABLED=no volumes: - "./:/home/jovyan/work:rw" ports: - '8081:8081' spark-nb: image: jupyter/pyspark-notebook:java-17.0.5 environment: - SPARK_MASTER_URL=spark://spark:7077 volumes: - "./:/home/jovyan/work:rw" ports: - '8888:8888' - '4040:4040'Run the command to deploy Docker Compose:
docker-compose upAccess the Jupyter notebook using the link logged in Docker Compose logs.
The Spark master URL is
spark://spark:7077.
# How do you read data stored in GCS on pandas with your local computer?
To do this:
Install
gcsfs:pip install gcsfsCopy the URI path to the file and use the following command to read it:
df = pandas.read_csv('gs://path/to/your/file.csv')
# TypeError when using spark.createDataFrame function on a pandas df
Error:
spark.createDataFrame(df_pandas).schema
TypeError: field Affiliated_base_number: Can not merge type <class 'pyspark.sql.types.StringType'> and <class 'pyspark.sql.types.DoubleType'>
Solution:
Reason:
- The
Affiliated_base_numberfield is a mix of letters and numbers, so it cannot be set toDoubleType. The suitable type would beStringType. Spark'sinferSchemais more accurate than Pandas' infer type method in this case. SetinferSchematotruewhen reading the CSV to prevent data removal.
- The
Implementation:
df = spark.read \ .options( header = "true", \ inferSchema = "true" ) \ .csv('path/to/your/csv/file/')Alternative Solution:
- Problem: Some rows in
affiliated_base_numberare null, and therefore are assigned the datatypeString, which cannot be converted toDouble. - Solution: Only take rows from the pandas df that are not null in the 'Affiliated_base_number' column before converting it to a PySpark DataFrame.
# Only take rows that have no null values pandas_df = pandas_df[pandas_df.notnull().all(1)]- Problem: Some rows in
# MemoryManager: Total allocation exceeds 95.00% (1,020,054,720 bytes) of heap memory
Default executor memory is 1GB. This error appeared when working with the homework dataset.
Error:
MemoryManager: Total allocation exceeds 95.00% (1,020,054,720 bytes) of heap memoryScaling row group sizes to 95.00% for 8 writers
Solution:
Increase the memory of the executor when creating the Spark session like this:
spark = SparkSession.builder \
.master("local[*]") \
.appName('test') \
.config("spark.executor.memory", "4g") \
.config("spark.driver.memory", "4g") \
.getOrCreate()
Remember to restart the Jupyter session (i.e., close the Spark session) or the config won’t take effect.
# How to spark standalone cluster is run on windows OS
Change the working directory to the Spark directory:
If you have set up your
SPARK_HOMEvariable, use the following:cd %SPARK_HOME%If not, use the following:
cd <path to spark installation>
Creating a Local Spark Cluster:
To start Spark Master:
bin\spark-class org.apache.spark.deploy.master.Master --host localhostStarting up a cluster:
bin\spark-class org.apache.spark.deploy.worker.Worker spark://localhost:7077 --host localhost
# Env variables set in ~/.bashrc are not loaded to Jupyter in VS Code
I added PYTHONPATH, JAVA_HOME, and SPARK_HOME to ~/.bashrc. Importing pyspark worked in iPython in the terminal but couldn't be found in a .ipynb file opened in VS Code.
After adding new lines to ~/.bashrc, you need to restart the shell to activate the changes. You can do either of the following:
source ~/.bashrc
or
exec bash
Instead of configuring paths in ~/.bashrc, you can create a .env file in the root of your workspace with the following content:
JAVA_HOME="${HOME}/app/java/jdk-11.0.2"
PATH="${JAVA_HOME}/bin:${PATH}"
SPARK_HOME="${HOME}/app/spark/spark-3.3.2-bin-hadoop3"
PATH="${SPARK_HOME}/bin:${PATH}"
PYTHONPATH="${SPARK_HOME}/python/:$PYTHONPATH"
PYTHONPATH="${SPARK_HOME}/python/lib/py4j-0.10.9.5-src.zip:$PYTHONPATH"
PYTHONPATH="${SPARK_HOME}/python/lib/pyspark.zip:$PYTHONPATH"
# hadoop “wc -l” is giving a different result than shown in the video
If you are using wc -l fhvhv_tripdata_2021-01.csv.gz with the gzip file as the file argument, you will get a different result, obviously, since the file is compressed.
Unzip the file and then use:
wc -l fhvhv_tripdata_2021-01.csv
to get the right results.
# Hadoop: Exception in thread "main" java.lang.UnsatisfiedLinkError: org.apache.hadoop.io.nativeio.NativeIO$Windows.access0(Ljava/lang/String;I)Z
If you are seeing this (or similar) error when attempting to write to parquet, it is likely an issue with your path variables.
For Windows, follow these steps:
- Create a new User Variable "HADOOP_HOME" that points to your Hadoop directory.
- Add "%HADOOP_HOME%\bin" to the PATH variable.
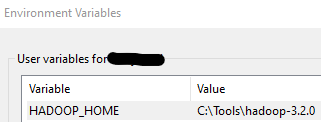
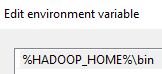
Additional tips can be found here: Stack Overflow
# Java.io.IOException. Cannot run program “C:\hadoop\bin\winutils.exe”. CreateProcess error=216, This version of 1% is not compatible with the version of Windows you are using.
To resolve the issue, follow these steps:
- Change the Hadoop version to 3.0.1.
- Replace all the files in the local Hadoop
binfolder with the files from this repository: winutils/hadoop-3.0.1/bin at master · cdarlint/winutils. - If this does not work, try other versions available in the repository.
For more information, refer to the following issue discussion: This version of %1 is not compatible with the version of Windows you're running · Issue #20 · cdarlint/winutils
# Dataproc: ERROR: (gcloud.dataproc.jobs.submit.pyspark) The required property [project] is not currently set. It can be set on a per-command basis by re-running your command with the [--project] flag.
Fix is to set the flag like the error states. Get your project ID from your dashboard and set it like so:
gcloud dataproc jobs submit pyspark \
--cluster=my_cluster \
--region=us-central1 \
--project=my-dtc-project-1010101 \
gs://my-dtc-bucket-id/code/06_spark_sql.py \
--
<{IMAGE:image_id}>
# Run Local Cluster Spark in Windows 10 with CMD
Go to
%SPARK_HOME%\binRun the following command to start the master:
spark-class org.apache.spark.deploy.master.MasterThis will give you a URL of the form
spark://ip:port.Run the following command to start the worker, replacing
spark://ip:portwith the URL obtained from the previous step:spark-class org.apache.spark.deploy.worker.Worker spark://ip:portCreate a new Jupyter notebook and set up the Spark session:
spark = SparkSession.builder \ .master("spark://{ip}:7077") \ .appName('test') \ .getOrCreate()Check on the Spark UI to see the master, worker, and application running.
# lServiceException: 401 Anonymous caller does not have storage.objects.list access to the Google Cloud Storage bucket. Permission 'storage.objects.list' denied on resource (or it may not exist).
This occurs because you are not logged in with Google Cloud SDK, or the project ID is not set. Follow these steps:
Log in using Google Cloud SDK:
gcloud auth loginThis will open a tab in the browser. Accept the terms, then close the tab if needed.
Set the project ID:
gcloud config set project <YOUR_PROJECT_ID>Upload the
pqdirectory to a Google Cloud Storage (GCS) Bucket:gsutil -m cp -r pq/ <YOUR_URI_from_gsutil>/pq
# GCP: py4j.protocol.Py4JJavaError
When submitting a job, you might encounter a py4j.protocol.Py4JJavaError related to Java in the log panel within Dataproc.
To address this error, consider the following steps:
Cluster Versioning Control:
- If you've recently changed the versioning settings, ensure that the cluster configuration is compatible with your requirements. For example, switching from Debian-Hadoop-Spark to Ubuntu 20.02-Hadoop3.3-Spark3.3 might resolve issues if you have a similar setup on your local machine.
Consistency with Local Environment:
- Aligning the cluster's OS version and software stack with your local environment can help reduce configuration issues.
Although specific documentation may not be available, this approach has proven effective in some scenarios.
# Repartition the Dataframe to 6 partitions using df.repartition(6) - got 8 partitions instead
Use both repartition and coalesce, like so:
df = df.repartition(6)
df = df.coalesce(6)
df.write.parquet('fhv/2019/10', mode='overwrite')
# Jupyter Notebook or SparkUI not loading properly at localhost after port forwarding from VS code?
Possible Solution: Try to forward the port using SSH CLI instead of VS Code.
Run the following command:
ssh -L <local port>:<VM host/ip>:<VM port> <ssh hostname>
ssh hostnameis the name you specified in the~/.ssh/configfile.
For example, in case of Jupyter Notebook, run:
ssh -L 8888:localhost:8888 gcp-vm
from your local machine’s CLI.
Note: If you logout from the session, the connection would break. While creating the Spark session, check the block's log because sometimes it fails to run at 4040 and switches to 4041.
If you are having trouble accessing localhost ports from a GCP VM, consider adding the forwarding instructions to the .ssh/config file as follows:
Host <hostname>
Hostname <external-gcp-ip>
User xxxx
IdentityFile yyyy
LocalForward 8888 localhost:8888
LocalForward 8080 localhost:8080
LocalForward 5432 localhost:5432
LocalForward 4040 localhost:4040
This should automatically forward all ports and will enable accessing localhost ports.
# Installing Java 11 on codespaces
Use the command below to check for available Java SDK versions:
sdk list javaInstall the desired version, for example:
sdk install java 11.0.22-amznIf prompted, press 'Y' to change the default Java version.
Verify the installation by checking the Java version:
java -versionIf the version does not work correctly, set the default version with:
sdk default java 11.0.22-amznAdjust this command to match the version you have installed.
# Error: Insufficient 'SSD_TOTAL_GB' quota. Requested 500.0, available 470.0.
Sometimes while creating a dataproc cluster on GCP, the following error is encountered.

Solutions:
As mentioned here, sometimes there might not be enough resources in the given region to allocate the request. Usually, resources get freed up in a bit, and one can create a cluster.
Changing the type of boot-disk from PD-Balanced to PD-Standard in Terraform helped solve the problem.
# Homework: how to convert the time difference of two timestamps to hours
Pyspark converts the difference of two TimestampType values to Python's native datetime.timedelta object. The timedelta object stores the duration in terms of days, seconds, and microseconds. Each of these units must be manually converted into hours to express the total duration between the two timestamps using only hours.
Another method to achieve this is using the datediff SQL function. It requires the following parameters:
- Upper Date: The closer date, e.g.,
dropoff_datetime. - Lower Date: The farther date, e.g.,
pickup_datetime.
The result is returned in days, so you can multiply the result by 24 to get the duration in hours.
# PicklingError: Could not serialize object: IndexError: tuple index out of range
This version combination worked for resolving the issue:
- PySpark: 3.3.2
- Pandas: 1.5.3
If you continue to encounter the error:
Py4JJavaError: An error occurred while calling o180.showString. : org.apache.spark.SparkException: Job aborted due to stage failure: Task 0 in stage 6.0 failed 1 times, most recent failure: Lost task 0.0 in stage 6.0 (TID 6) (host.docker.internal executor driver): org.apache.spark.SparkException: Python worker failed to connect back.
Try running the following before initializing SparkSession:
import os
import sys
os.environ['PYSPARK_PYTHON'] = sys.executable
os.environ['PYSPARK_DRIVER_PYTHON'] = sys.executable
# RuntimeError: Python in worker has different version 3.11 than that in driver 3.10, PySpark cannot run with different minor versions. Please check environment variables PYSPARK_PYTHON and PYSPARK_DRIVER_PYTHON are correctly set.
To resolve the version mismatch error between the worker and driver Python versions in PySpark, set the environment variables PYSPARK_PYTHON and PYSPARK_DRIVER_PYTHON to the same executable.
import os
import sys
os.environ['PYSPARK_PYTHON'] = sys.executable
os.environ['PYSPARK_DRIVER_PYTHON'] = sys.executable
For further information on Dataproc Pricing, visit: Dataproc Pricing
# Dataproc Qn: Is it essential to have a VM on GCP for running Dataproc and submitting jobs?
No, you can submit a job to Dataproc from your local computer by installing and configuring gsutil. For installation instructions, visit https://cloud.google.com/storage/docs/gsutil_install.
You can execute the following command from your local computer:
gcloud dataproc jobs submit pyspark \
--cluster=de-zoomcamp-cluster \
--region=europe-west6 \
gs://dtc_data_lake_de-zoomcamp-nytaxi/code/06_spark_sql.py \
-- \
--input_green=gs://dtc_data_lake_de-zoomcamp-nytaxi/pq/green/2020/*/ \
--input_yellow=gs://dtc_data_lake_de-zoomcamp-nytaxi/pq/yellow/2020/*/ \
--output=gs://dtc_data_lake_de-zoomcamp-nytaxi/report-2020
# In module 5.3.1, trying to run `spark.createDataFrame(df_pandas).show()` returns error
AttributeError: 'DataFrame' object has no attribute 'iteritems'
This error occurs because a method inside PySpark refers to a package that has been deprecated (Stack Overflow).
Solutions
Refer to the code mentioned in the Stack Overflow link.
Another workaround is to create a conda environment to downgrade Python's version to 3.8 and Pandas to 1.5.3:
conda create -n pyspark_env python=3.8 pandas=1.5.3 jupyter conda activate pyspark_env
# Cannot create a cluster: Insufficient 'SSD_TOTAL_GB' quota. Requested 500.0, available 250.0.
The master and worker nodes are allocated a maximum of 250 GB of memory combined. In the configuration section, adhere to the following specifications:
Master Node:
- Machine type:
n2-standard-2 - Primary disk size: 85 GB
- Machine type:
Worker Node:
- Number of worker nodes: 2
- Machine type:
n2-standard-2 - Primary disk size: 80 GB
You can allocate up to 82.5 GB memory for worker nodes, keeping in mind that the total memory allocated across all nodes cannot exceed 250 GB.
# Setting JAVA_HOME with Homebrew on Apple Silicon
The MacOS setup instruction (here) for setting the JAVA_HOME environment variable is for Intel-based Macs, which have a default install location at /usr/local/. If you have an Apple Silicon Mac, you need to set JAVA_HOME to /opt/homebrew/. Update your .bashrc or .zshrc file with the following:
export JAVA_HOME="/opt/homebrew/opt/openjdk/bin"
export PATH="$JAVA_HOME:$PATH"
Confirm that your path was correctly set by running the command:
which java
You should expect to see the output:
/opt/homebrew/opt/openjdk/bin/java
Check the Java version with the following command:
java -version
Reference: https://docs.brew.sh/Installation
# Subnetwork 'default' does not support Private Google Access which is required for Dataproc clusters when 'internal_ip_only' is set to 'true'. Enable Private Google Access on subnetwork 'default' or set 'internal_ip_only' to 'false'.
To resolve this issue, follow these steps:
- Search for VPC in Google Cloud Console.
- Navigate to the "SUBNETS IN CURRENT PROJECT" tab.
- Locate the region/location where your Dataproc will be located and click on it.
- Click the edit button and toggle on "Private Google Access."
- Save changes.
Module 6: Streaming with Kafka
# Spark: Is working, however, nothing appears in the Spark UI (e.g., .show())?
Since we used multiple notebooks during the course, it's possible that there are more than one Spark session active. It’s highly likely that you are observing the incorrect one. Follow these steps to troubleshoot:
Spark uses port 4040 by default, but if more than one session is active, it will try to use the next available port (e.g., 4041).
Ensure you're viewing the correct Spark Web UI for the application where your jobs are running.
Verify the current application session address:
# Using the following command in your session spark.sparkContext.uiWebUrlExpected output might look like:
http://your.application.session.address.internal:4041Indicating port 4041.
If using a VM, make sure you forward the identified port to access the web UI on
localhost:<port>.
# Docker: Could not start docker image "control-center" from the docker-compose.yaml file.
Check Docker Compose File:
Ensure that your docker-compose.yaml file is correctly configured with the necessary details for the "control-center" service. Check the service name, image name, ports, volumes, environment variables, and any other configurations required for the container to start.
On Mac OSX 12.2.1 (Monterey), if you encounter this issue, try the following:
- Open Docker Desktop and check for any images still running from previous sessions.
- If there are images still running and they do not appear with the
docker pscommand, they may need to be deleted directly from Docker Desktop. - Once those images are removed, try starting the Kafka environment again.
This approach resolved the issue on Mac OSX 12.2.1 for a similar setup.
# Module “kafka” not found when trying to run producer.py
To resolve the "Module 'kafka' not found" error when running producer.py, you can create a virtual environment and install the required packages. Follow these steps:
Create a Virtual Environment
Run the following command to create a virtual environment:
python -m venv envActivate the Virtual Environment
On macOS and Linux:
source env/bin/activateOn Windows:
env\Scripts\activate
Install Required Packages
Install the packages listed in
requirements.txt:pip install -r ../requirements.txtDeactivate the Virtual Environment
When you're done, deactivate the virtual environment:
deactivate
Note: Ensure that Docker images are running before executing the Python file. The virtual environment is meant for running the Python files locally.
# Error: Importing cimpl DLL when running Avro examples
ImportError: DLL load failed while importing cimpl: The specified module could not be found
Steps to Resolve:
Verify Python Version:
Ensure you are using a compatible version of Python with the Avro library. Check the Python version and compatibility requirements specified by the Avro library documentation.
Load Required DLL:
You may need to load
librdkafka-5d2e2910.dllin your code before importing Avro. Add the following:from ctypes import CDLL CDLL("C:\\Users\\YOUR_USER_NAME\\anaconda3\\envs\\dtcde\\Lib\\site-packages\\confluent_kafka.libs\\librdkafka-5d2e2910.dll")Note that this error may occur depending on the OS and Python version installed.
Alternative Solution:
If you encounter
ImportError: DLL load failed while importing cimpl, you can try the following solution in PowerShell:$env:CONDA_DLL_SEARCH_MODIFICATION_ENABLE=1This sets the DLL manually in the Conda environment.
Source:
# ModuleNotFoundError: No module named 'avro'
Solution
To resolve the error, install the Avro module using the following command:
pip install confluent-kafka[avro]
Note: This issue may occur because Conda does not include the Avro module when installing confluent-kafka via pip.
Additional Resources
For more information on Anaconda and confluent-kafka issues, visit the following links:
# Error while running python3 stream.py worker
If you get an error while running the command python3 stream.py worker, follow these steps:
Uninstall the current
kafka-pythonpackage:pip uninstall kafka-pythonInstall the specific version of
kafka-python:pip install kafka-python==1.4.6
# What is the use of Redpanda?
Redpanda is built on top of the Raft consensus algorithm and is designed as a high-performance, low-latency alternative to Kafka. It uses a log-centric architecture similar to Kafka but with different underlying principles.
# Negsignal:SIGKILL while converting data files to parquet format
Got this error because the Docker container memory was exhausted. The data file was up to 800MB but my Docker container does not have enough memory to handle that.
Solution:
- Load the file in chunks with Pandas.
- Create multiple parquet files for each data file being processed.
This approach worked smoothly and resolved the issue.
# resources/rides.csv is missing
Copy the file found in the Java example: data-engineering-zoomcamp/week_6_stream_processing/java/kafka_examples/src/main/resources/rides.csv
# Kafka: Python videos have low audio and are hard to follow up
To improve the audio quality:
- Download the videos and use VLC media player. You can set the audio to 200% of the original volume for better sound quality.
- Alternatively, use auto-generated captions directly on YouTube for better clarity.
# Kafka Python Videos: Rides.csv
There is no clear explanation of the rides.csv data that the producer.py Python programs use. You can find it here: Rides CSV File.
# kafka.errors.NoBrokersAvailable: NoBrokersAvailable
If you encounter this error, it is likely that your Kafka broker Docker container is not running.
Use the following command to check the running containers:
docker psNavigate to the folder containing your Docker Compose YAML file and execute the following command to start all instances:
docker-compose up -d
# Kafka homework Q3: There are options that support the scaling concept more than the others.
Focus on the horizontal scaling option.
Think of scaling in terms of scaling from the consumer end, or consuming messages via horizontal scaling.
# Docker: How to fix docker compose error: Error response from daemon: pull access denied for spark-3.3.1, repository does not exist or may require 'docker login': denied: requested access to the resource is denied
If you get this error, it means you have not built your Spark and Jupyter images. These images aren’t readily available on DockerHub.
To resolve this:
In the Spark folder, run the following command from a bash CLI to build all images before running docker compose:
./build.sh
# Python Kafka: ./build.sh: Permission denied Error
Run this command in the terminal in the same directory (/docker/spark):
chmod +x build.sh
# Python Kafka: ‘KafkaTimeoutError: Failed to update metadata after 60.0 secs.’ when running stream-example/producer.py
Restarting all services worked for me:
docker-compose down
docker-compose up
# Python Kafka: ./spark-submit.sh streaming.py - ERROR StandaloneSchedulerBackend: Application has been killed. Reason: All masters are unresponsive! Giving up.
While following tutorial 13.2, when running ./spark-submit.sh streaming.py, encountered the following error:
24/03/11 09:48:36 INFO StandaloneAppClient$ClientEndpoint: Connecting to master spark://localhost:7077...
24/03/11 09:48:36 INFO TransportClientFactory: Successfully created connection to localhost/127.0.0.1:7077 after 10 ms (0 ms spent in bootstraps)
24/03/11 09:48:54 WARN GarbageCollectionMetrics: To enable non-built-in garbage collector(s) List(G1 Concurrent GC), users should configure it(them) to spark.eventLog.gcMetrics.youngGenerationGarbageCollectors or spark.eventLog.gcMetrics.oldGenerationGarbageCollectors
24/03/11 09:48:56 INFO StandaloneAppClient$ClientEndpoint: Connecting to master spark://localhost:7077…
24/03/11 09:49:16 INFO StandaloneAppClient$ClientEndpoint: Connecting to master spark://localhost:7077...
24/03/11 09:49:36 WARN StandaloneSchedulerBackend: Application ID is not initialized yet.
24/03/11 09:49:36 ERROR StandaloneSchedulerBackend: Application has been killed. Reason: All masters are unresponsive! Giving up.
py4j.protocol.Py4JJavaError: An error occurred while calling None.org.apache.spark.sql.SparkSession.
: java.lang.IllegalStateException: Cannot call methods on a stopped SparkContext.
Solution:
Downgrade PySpark:
- Downgrade your local PySpark to 3.3.1 (ensure it matches the version used in Dockerfile).
- The mismatch of PySpark versions can be a cause of the failed connection.
- Check the logs of
spark-masterin the Docker container for confirmation.
Check Spark Version:
- Run
pyspark --versionandspark-submit --versionto check your local Spark version. - Adjust the
SPARK_VERSIONvariable inbuild.shto match your current Spark version.
- Run
# Python Kafka: ./spark-submit.sh streaming.py - How to check why Spark master connection fails
Start a new terminal.
Run the following command to list running containers:
docker psCopy the
CONTAINER IDof thespark-mastercontainer.Execute the following command to access the container's shell:
docker exec -it <spark_master_container_id> bashRun this command to view the Spark master logs:
cat logs/spark-master.outCheck the log for the timestamp when the error occurred.
Search the error message online for further troubleshooting.
# Python Kafka: ./spark-submit.sh streaming.py Error: py4j.protocol.Py4JJavaError: An error occurred while calling None.org.apache.spark.api.java.JavaSparkContext.
Make sure your Java version is 11 or 8.
Check your version by:
java --versionCheck all your installed Java versions by:
/usr/libexec/java_home -VIf you already have Java 11 but it's not set as the default, select the specific version by:
export JAVA_HOME=$(/usr/libexec/java_home -v 11.0.22)(or another version of 11)
# Java Kafka: <project_name>-1.0-SNAPSHOT.jar errors: package xxx does not exist even after gradle build
In my setup, all of the dependencies listed in build.gradle were not installed in <project_name>-1.0-SNAPSHOT.jar.
Solution:
In the
build.gradlefile, add the following at the end:shadowJar { archiveBaseName = "java-kafka-rides" archiveClassifier = '' }In the command line, run:
gradle shadowjarExecute the script from
java-kafka-rides-1.0-SNAPSHOT.jarcreated by the shadowjar.
# Python Kafka: Installing dependencies for python3 06-streaming/python/avro_example/producer.py
To install the necessary dependencies for running producer.py in the avro_example directory, use the following commands:
Install
confluent-kafka:- Using pip:
pip install confluent-kafka- Using conda:
conda install conda-forge::python-confluent-kafkaInstall
fastavro:pip install fastavro
# Can I install the Faust Library for Module 6 Python Version due to dependency conflicts?
The Faust repository and library is no longer maintained - GitHub Repository
If you do not know Java, you now have the option to follow the Python Videos 6.13 & 6.14 here: YouTube Playlist. Follow the RedPanda Python version here: RedPanda Example.
Note: I highly recommend watching the Java videos to understand the concept of streaming, but you can skip the coding parts. All will become clear when you get to the Python videos and RedPanda files.
# Java Kafka: How to run producer/consumer/kstreams/etc in terminal
In the project directory, run:
java -cp build/libs/<jar_name>-1.0-SNAPSHOT.jar:out src/main/java/org/example/JsonProducer.java
# Java Kafka: When running the producer/consumer/etc java scripts, no results retrieved or no message sent
For example, when running JsonConsumer.java, you might see:
Consuming form kafka started
RESULTS:::0
RESULTS:::0
RESULTS:::0
Or when running JsonProducer.java, you might encounter:
Exception in thread "main" java.util.concurrent.ExecutionException: org.apache.kafka.common.errors.SaslAuthenticationException: Authentication failed
Solution:
Ensure the
StreamsConfig.BOOTSTRAP_SERVERS_CONFIGin the scripts located atsrc/main/java/org/example/(e.g.,JsonConsumer.java,JsonProducer.java) is pointing to the correct server URL (e.g.,europe-west3vseurope-west2).Verify that the cluster key and secrets are updated in
src/main/java/org/example/Secrets.java(KAFKA_CLUSTER_KEYandKAFKA_CLUSTER_SECRET).
# Java Kafka: Tests are not picked up in VSCode
In VS Code, you might expect to see a triangle icon next to each test method in your Java files. If you don't see it, here are the steps to fix the issue:
Open Explorer (first icon on the left navigation bar).
Navigate to JAVA PROJECTS (bottom collapsable section).
Click the icon in the rightmost position next to JAVA PROJECTS to open the options.


Select Clean Workspace.
Confirm by clicking Reload and Delete.
Following these steps should restore the triangle icons you expect to see next to each test, similar to those visible in Python tests.
Example:
Additionally, you can add classes and packages in this window instead of creating files directly in the project directory.
# Confluent Kafka: Where can I find schema registry URL?
In Confluent Cloud:
- Navigate to your Environment (e.g., default or a custom name).
- Use the right navigation bar to find "Stream Governance API."
- The URL can be found under "Endpoint."
- Create credentials from the Credentials section below it.
# How do I check compatibility of local and container Spark versions?
You can check the version of your local Spark using:
spark-submit --version
In the build.sh file of the Python folder, ensure that SPARK_VERSION matches your local version. Similarly, ensure the PySpark you installed via pip also matches this version.
# How to fix the error "ModuleNotFoundError: No module named 'kafka.vendor.six.moves'?"
# How to fix “connection failed: connection to server at "127.0.0.1", port 5432 failed” error when setting up Postgres connection in pgAdmin?
Instead of using “localhost” as the host name/address, try “postgres”, or “host.docker.internal” instead.
Alternative Solution:
If you have installed Postgres locally and disabled persist data on the Postgres container in Docker (i.e., volume: removed), use a Postgres port other than 5432, such as 5433.
For the pgAdmin host name/address, if 'localhost', 'postgres', or 'host.docker.internal' are not working, you can use your own IPv4 Address.
To find your IPv4 Address on Windows OS:
Open Command Prompt.
Run the command:
ipconfigLook under Wireless LAN adapter WiFi 2 for the IPv4 Address. For example:
IPv4 Address. . . . . . . . . . . : 192.168.0.148
Project
# Why is my table not being created in PostgreSQL when I submit a job?
There could be a few reasons for this issue:
Race Conditions: If you're running multiple processes in parallel.
Database Connection Issues: The job might not be connecting to the correct PostgreSQL database, or there could be authentication or permission issues preventing table creation.
Missing Table Creation Logic: The code responsible for creating the table might not be properly included or executed in the job submission process.
As a best practice, it's generally recommended to pre-create tables in PostgreSQL to avoid runtime errors. This ensures the database schema is properly set up before any jobs are executed.
Extra: Use CREATE TABLE IF NOT EXISTS in your code. This will prevent errors if the table already exists and ensure smooth job execution.
# How is my capstone project going to be evaluated?
Each submitted project will be evaluated by three randomly assigned students who have also submitted the project.
You will also be responsible for grading projects from three fellow students yourself. Please note that not complying with this rule will result in failing to achieve the Certificate at the end of the course.
The final grade you receive will be the median score of the grades from peer reviewers.
The peer review criteria for evaluating or being evaluated must follow the guidelines defined here.
# Can I collaborate with others on the capstone project?
Collaboration is not allowed for the capstone submission. However, you can discuss ideas and get feedback from peers in the forums or Slack channels.
# Project 1 & Project 2
There is only ONE project for this Zoomcamp. You do not need to submit or create two projects.
There are simply TWO chances to pass the course. You can use the Second Attempt if you:
- Fail the first attempt
- Do not have the time due to other engagements such as holidays or sickness to enter your project into the first attempt.
Project Evaluation - Reproducibility
Even with thorough documentation, ensuring that a peer reviewer can follow your steps can be challenging. Here’s how this criterion will be evaluated:
"Ideally yes, you should try to re-run everything. But I understand that not everyone has time to do it, so if you check the code by looking at it and try to spot errors, places with missing instructions, and so on - then it's already great."
Certificates: How do I get it?
See the certificate.mdx file.
# Does anyone know nice and relatively large datasets?
See a list of datasets here: GitHub Datasets
# How to run Python as a startup script?
You need to redefine the Python environment variable to that of your user account.
# Spark Streaming: How do I read from multiple topics in the same Spark Session
To read from multiple topics in the same Spark session, follow these steps:
Initiate a Spark Session:
spark = (SparkSession .builder .appName(app_name) .master(master=master) .getOrCreate()) spark.streams.resetTerminated()Read Streams from Multiple Topics:
query1 = spark .readStream ... ... .load() query2 = spark .readStream ... ... .load() query3 = spark .readStream ... ... .load()Start the Queries:
query1.start() query2.start() query3.start()Await Termination:
spark.streams.awaitAnyTermination() # Waits for any one of the queries to receive a kill signal or error failure. This is asynchronous.Note:
query3.start().awaitTermination()is a blocking call. It works well when we are reading only from one topic.
# Data Transformation from Databricks to Azure SQL DB
Transformed data can be moved into Azure Blob Storage and then it can be moved into Azure SQL DB, instead of moving directly from Databricks to Azure SQL DB.
# Orchestrating dbt with Airflow
The trial dbt account provides access to the dbt API. A job will still need to be added manually. Airflow can run the job using a Python operator that calls the API. You will need to provide an API key, job ID, etc., and be careful not to commit this information to GitHub.
- Detailed explanation: dbt and Airflow Spiritual Alignment
- Source code example: GitHub dbt Cloud Example
# Orchestrating DataProc with Airflow
For orchestrating DataProc with Airflow, you can refer to the following documentation:
Roles for Service Account
Ensure that you assign the following roles to your service account:
DataProc Administrator
Service Account User
For more details, see the explanation on Stack Overflow.
Operators to Use
DataprocSubmitPySparkJobOperatorDataprocDeleteClusterOperatorDataprocCreateClusterOperator
Important Note
When using DataprocSubmitPySparkJobOperator, make sure to add the BigQuery Connector, as DataProc does not include it by default:
dataproc_jars = ["gs://spark-lib/bigquery/spark-bigquery-with-dependencies_2.12-0.24.0.jar"]
# Orchestrating dbt cloud with Mage
You can trigger your dbt job in a Mage pipeline. Follow these steps:
Retrieve your dbt cloud API key from Settings > API Tokens > Personal Tokens. Add this key safely to your
.envfile. For example:dbt_api_trigger=dbt_**Navigate to the job page in dbt cloud and find the API trigger link.
Create a custom Mage Python block with a simple HTTP request, as shown in this example.

Use the following script to trigger the dbt job:
from dotenv import load_dotenv from pathlib import Path dotenv_path = Path('/home/src/.env') load_dotenv(dotenv_path=dotenv_path) dbt_api_trigger = os.getenv('dbt_api_trigger') url = f"https://cloud.getdbt.com/api/v2/accounts/{dbt_account_id}/jobs/<job_id>/run/" headers = { "Authorization": f"Token {dbt_api_trigger}", "Content-Type": "application/json" } body = { "cause": "Triggered via API" } response = requests.post(url, headers=headers, json=body)Voilà! You've triggered a dbt job from your Mage pipeline.
# Key Vault in Azure cloud stack
The Key Vault in Azure Cloud is used to store credentials, passwords, or secrets for different technologies used within Azure. For example, if you do not want to expose the password of an SQL database, you can save the password under a given name and use it in other Azure services.
# How to connect Pyspark with BigQuery?
To connect Pyspark with BigQuery, include the following line in the Pyspark configuration:
# Example initialization of SparkSession variable
spark = (SparkSession.builder
.master("...")
.appName("...")
# Add the following configuration
.config("spark.jars.packages", "com.google.cloud.spark:spark-3.5-bigquery:0.37.0")
)
# How to run a dbt-core project as an Airflow Task Group on Google Cloud Composer using a service account JSON key
Install the astronomer-cosmos package as a dependency. Refer to the installation guide here and see a Terraform example.
Create a new folder,
dbt/, inside thedags/folder of your Composer GCP bucket and copy your dbt-core project there. See the example.Ensure your
profiles.ymlis configured to authenticate with a service account key. Refer to the BigQuery example.Create a new DAG using the
DbtTaskGroupclass. Use aProfileConfigspecifying aprofiles_yml_filepaththat points to the location of your JSON key file. See this example.
Your dbt lineage graph should now appear as tasks inside a task group like this:
# How can I run UV in Kestra without installing it on every flow execution?
To avoid reinstalling uv on each flow run, you can create a custom Docker image based on the official Kestra image with uv pre-installed. Here's how:
Create a Dockerfile (e.g., Dockerfile) with the following content:
# Use the official Kestra image as a base FROM kestra/kestra # Install uv RUN pip install uvUpdate your
docker-compose.ymlto build this custom image instead of pulling the default one:services: kestra: build: context: . dockerfile: Dockerfile image: custom-kestra
This approach ensures that uv is available in the container at runtime without requiring installation during each flow execution.
# Is it possible to create external tables in BigQuery using URLs, such as those from the NY Taxi data website?
Not really, only Bigtable, Cloud Storage, and Google Drive are supported data stores.
# Is it ok to use NY_Taxi data for the project?
No.
# How to use dbt-core with Athena?
If you don’t have access to dbt Cloud, which is natively supported by AWS, you can use the community-built dbt-Athena Adapter for dbt-Core. Here are some references:
Key Features:
- Enables dbt to work with AWS Athena using dbt Core
- Allows data transformation using
CREATE TABLE ASorCREATE VIEWSQL queries
Not Yet Supported Features:
- Python models
- Persisting documentation for views
This adapter can be a valuable resource for those who need to work with Athena using dbt Core.
Workshop 1 - dlthub
# Solving dbt-Athena library conflicts
When working on a dbt-Athena project, do not install dbt-athena-adapter. Instead, always use the dbt-athena-community package, ensuring it matches the version of dbt-core to avoid compatibility conflicts.
Best Practice
Always pin the versions of
dbt-coreanddbt-athena-communityto the same version.Example: dbt-core~=1.9.3 dbt-athena-community~=1.9.3
Why?
dbt-athena-adapteris outdated and no longer maintained.dbt-athena-communityis the actively maintained package and is compatible with the latest versions ofdbt-core.
Steps to Avoid Conflicts
- Check the compatibility matrix in the dbt-athena-community GitHub repository.
- Update
requirements.txtto use the latest compatible versions ofdbt-coreanddbt-athena-community. - Avoid mixing
dbt-athena-adapterwithdbt-athena-communityin the same environment.
By following this practice, you can avoid the conflicts we faced previously and ensure a smooth development experience.
# Which set-up should I use for my dlt homework?
Technically you can use any code editor or Jupyter Notebook, as long as you can run dbt and answer the homework questions. A lot of code is provided by the instructor on the homework page to give you a head start in the right direction: dlt Homework Instructions.
The most practical way is to use the provided Colabs Jupyter notebook called ‘dlt - Homework.ipynb’ which you can find here: Colab Notebook since all of the provided code is applicable in the Colabs set-up.
# How do I install the necessary dependencies to run the code?
To run the provided code, ensure that the dlt[duckdb] package is installed. You can do this by executing the following installation command in a Jupyter notebook:
!pip install dlt[duckdb]
If you’re installing it locally, make sure to also have duckdb installed before the duckdb package is loaded:
pip install "dlt[duckdb]"
# Other packages needed but not listed
If you are running Jupyter Notebook on a fresh new Codespace or in a local machine with a new virtual environment, you will need these packages to run the starter Jupyter Notebook offered by the teacher. Execute this command to install all the necessary dependencies:
pip install duckdb pandas numpy pyarrow
Or save it into a requirements.txt file:
dlt[duckdb]
duckdb
pandas
numpy
pyarrow # Optional, needed for Parquet support
Then run:
pip install -r requirements.txt
# How can I use DuckDB In-Memory database with dlt?
Alternatively, you can switch to in-file storage with:
# Homework: dlt Exercise 3 - Merge a generator concerns
After loading, you should have a total of 8 records, and ID 3 should have age 33.
Question: Calculate the sum of ages of all the people loaded as described above.
- The sum of all eight records' respective ages is too big to be in the choices.
- You need to first filter out the people whose occupation is equal to
Nonein order to get an answer that is close to or present in the given choices.
Issue:
I'm having an issue with the DLT workshop notebook, specifically in the 'Load to Parquet file' section. No matter what I change the file path to, it's still saving the DLT files directly to my C drive.
Solution:
Use a raw string and keep the file:/// at the start of your file path.
# Set the bucket_url. We can also use a local folder
os.environ['DESTINATION__FILESYSTEM__BUCKET_URL'] = r'file:///content/.dlt/my_folder'
url = "https://storage.googleapis.com/dtc_zoomcamp_api/yellow_tripdata_2009-06.jsonl"
# Define your pipeline
pipeline = dlt.pipeline(
pipeline_name='my_pipeline',
destination='filesystem',
dataset_name='mydata'
)
# Run the pipeline with the generator we created earlier.
load_info = pipeline.run(stream_download_jsonl(url), table_name="users", loader_file_format="parquet")
print(load_info)
# Get a list of all Parquet files in the specified folder
parquet_files = glob.glob('/content/.dlt/my_folder/mydata/users/*.parquet')
# Show Parquet files
for file in parquet_files:
print(file)



# Problem with importing the dlt or dlt.sources module
Make sure you don’t have a dlt.py file saved in the same directory as your working file.
# How to set credentials in Google Colab notebook to connect to BigQuery
In the secrets sidebar, create a secret BIGQUERY_CREDENTIALS with the value being your Google Cloud service account key. Then load it with:
import os
from google.colab import userdata
os.environ["DESTINATION__BIGQUERY__CREDENTIALS"] = userdata.get('BIGQUERY_CREDENTIALS')
# How do I set up credentials to run dlt in my environment (not Google Colab)?
You can set up credentials for dlt in several ways. Here are the two most common methods:
Environment Variables (Easiest)
Set credentials via environment variables. For example, to configure Google Cloud credentials. This method avoids hardcoding secrets in your code and works seamlessly with most environments.
Configuration Files (Recommended for Local Use)
- Use
.dlt/secrets.tomlfor sensitive credentials and.dlt/config.tomlfor non-sensitive configurations. - Example for Google Cloud in
secrets.toml:
[google_cloud]
service_account_key = "YOUR_SERVICE_ACCOUNT_KEY"
- Place these files in the
.dltfolder of your project.
Additional Notes:
- Never commit
secrets.tomlto version control (add it to.gitignore). - Credentials can also be loaded via vaults, AWS Parameter Store, or custom setups.
For additional methods and detailed information, refer to the official dlt documentation
# Make DLT comply with the XDG Base Dir Specification
You can set the environment variable in your shell init script:
For Bash or ZSH:
export DLT_DATA_DIR=$XDG_DATA_HOME/dlt
For Fish (in config.fish):
set -x DLT_DATA_DIR "$XDG_DATA_HOME/dlt"
# Embedding dlt into Apache Airflow
To integrate a dlt pipeline into Apache Airflow, follow this example:
from airflow import DAG
from airflow.operators.python import PythonOperator
from datetime import datetime, timedelta
import dlt
from my_dlt_pipeline import load_data # Import your dlt pipeline function
default_args = {
"owner": "airflow",
"depends_on_past": False,
"start_date": datetime(2024, 2, 16),
"retries": 1,
"retry_delay": timedelta(minutes=5),
}
def run_dlt_pipeline():
pipeline = dlt.pipeline(
pipeline_name="my_pipeline",
destination="duckdb", # Change this based on your database
dataset_name="my_dataset"
)
info = pipeline.run(load_data())
print(info) # Logs for debugging
with DAG(
"dlt_airflow_pipeline",
default_args=default_args,
schedule_interval="@daily",
catchup=False,
) as dag:
run_dlt_task = PythonOperator(
task_id="run_dlt_pipeline",
python_callable=run_dlt_pipeline,
)
run_dlt_task
Ensure to replace "duckdb" with your actual database name and adjust the load_data function according to your specific dlt pipeline.
# Embedding dlt into Kestra
id: dlt_ingestion
namespace: my.dlt
description: "Run dlt pipeline with Kestra"
tasks:
- id: run_dlt
type: io.kestra.plugin.scripts.python.Commands
commands:
- |
import dlt
from my_dlt_pipeline import load_data # Import your dlt function
pipeline = dlt.pipeline(
pipeline_name="kestra_pipeline",
destination="duckdb",
dataset_name="kestra_dataset"
)
info = pipeline.run(load_data())
print(info)
# Loading Dlt Exports from GCS Filesystems
When using the filesystem destination, you may have issues reading the files exported because DLT will by default compress the files. If you are using loader_file_format="parquet" then BigQuery should cope with this compression OK. If you want to use JSONL or CSV format, however, you may need to disable file compression to avoid issues with reading the files directly in BigQuery. To do this, set the following config:
[normalize.data_writer]
disable_compression = true
There is further information at DLTHub Docs.
Warning:
Test 'test.taxi_rides_ny.relationships_stg_yellow_tripdata_dropoff_locationid__locationid__ref_taxi_zone_lookup_csv_.085c4830e7' (models/staging/schema.yml) depends on a node named 'taxi_zone_lookup.csv' in package '' which was not found
Solution: This warning indicates that dbt is trying to reference a model or source named taxi_zone_lookup.csv, but it cannot find it. We might have a typo in our ref() function.
Tests:
Name: relationships_stg_yellow_tripdata_dropoff_locationid
Description: Ensure
dropoff_location_idexists intaxi_zone_lookup.csvRelationships:
To:
ref('taxi_zone_lookup.csv')# ❌ Wrong referenceField:
locationidTo:
ref('taxi_zone_lookup')# ✅ Correct reference
Pandas and Spark Version Mismatch:
When running df_spark = spark.createDataFrame(df_pandas), an error indicating a version mismatch between Pandas and Spark was encountered. To resolve this, either:
- Downgrade Pandas to a version below 2.
- Upgrade Spark to version 3.5.5.
I chose to upgrade Spark to 3.5.5, and it worked.
Avoiding Backpressure in Flink:
What’s Backpressure?
- It occurs when Flink processes data slower than Kafka produces it.
- This leads to increased memory usage and can slow down or crash the job.
How to Fix It?
- Adjust Kafka’s consumer parallelism to match the producer rate.
- Increase partitions in Kafka to allow more parallel processing.
- Monitor Flink metrics to detect backpressure.
env.set_parallelism(4) # Adjust parallelism to avoid bottlenecks